Impact of inbreeding on growth and development of young open-pollinated progeny of Eucalyptus globulus
iForest - Biogeosciences and Forestry, Volume 15, Issue 5, Pages 356-362 (2022)
doi: https://doi.org/10.3832/ifor4012-015
Published: Sep 20, 2022 - Copyright © 2022 SISEF
Research Articles
Abstract
The use of open-pollinated seeds from seed orchards is a common strategy for the deployment of genetically improved eucalypts, including Eucalyptus globulus, an important pulpwood tree in many temperate climate areas. However, seed quality can be affected by the rate of selfing and to a lesser extent by contamination from pollen outside the orchard. Inbreeding between related parents and especially from self-crosses is known to cause diminished growth and developmental abnormalities in the resulting progeny. This study looks at the magnitude and variation in selfing and the impact in inbreeding depression across several E. globulus families collected over the years in a seed orchard. The effects on growth and development of outcrossed and selfed progeny were studied across five progeny trials, after pedigree reconstruction of the open pollinated progeny based on SSR genotyping. An additive genetic mixed linear model was fitted to the data to evaluate the impact of inbreeding on height growth. The results showed a significant inbreeding depression, with a height growth reduction of 15% in selfed progeny, when compared with crosses from unrelated parents. These inbreeding depression values varied among families, ranging between 7% and 24%, evidencing the importance of genetic background. Contamination rates were on average 10% suggesting long distance pollen dispersal was present. A small number of abnormal phenotypes (less than 10%) was observed in the field. This was associated with specific, unrelated, crosses and not to high inbreeding rates such as found among selfed progeny. The relevance of these results for orchard management and parent selection is discussed.
Keywords
Inbreeding Depression, Seed Orchard, Selfing, Abnormal Phenotypes, SSRs, Pedigree Reconstruction
Introduction
Eucalyptus globulus Labill. is a tree species belonging to the Myrtaceae family, native to Tasmania and Victoria, in mainland Australia ([28]). It has been planted around the world due to its excellent growth and pulpwood characteristics ([1]). Introduced in Portugal in the 19th century, E. globulus currently covers around 850.000 ha of plantations supplying the local industry with nearly 6 million cubic meters per year of high-quality pulpwood ([8]).
In Portugal, breeding programs for this species have been in place for over 40 years, relying mostly on clonal and controlled crossing schemes ([3]). However, due to their simplicity and low costs, large-scale production of improved stocks has also been based on open pollinated seed orchards ([6], [21], [23]). Such system however has its own caveats. Although the species is mostly outcrossing and several self-incompatibility mechanisms have been reported ([42]), self-pollination is common ([41]). This can increase average co-ancestry and the concomitant build-up of inbreeding and inbreeding depression in the seed, resulting in losses of growth, survival and other adverse developmental consequences ([24], [11], [15], [19], [39], [43]). These adverse effects however are known to vary among genotypes ([36], [14]). Also, selfing rates can vary due to asynchronous flowering among the parents in the orchard or to low activity of pollinators ([40], [25], [26], [37]). Another factor influencing the genetic quality of seeds from seed orchards is the degree of contamination from unimproved pollen from outside the orchard, which depends on the degree of its spatial isolation. Reported contamination rates have ranged between 5% and 15% ([21], [43]) in isolated orchards but can reach 40% in smaller or less isolated locations ([9], [10], [44]). Thus, the amount of selfing and contamination from outside pollen in seed orchards are important seed quality parameters to quantify, monitor and if possible, to control. Finally, abnormal phenotypes, often observed in the field trials and plantations, have been previously associated with possible high levels of inbreeding, and particularly with selfing. This is expected since selfed plants (i.e., more inbred) tend to express more often, putative deleterious or prejudicial recessive alleles ([31], [19]). In E. grandis, similar abnormalities, including different leaf shape and excessive lateral branching, have similarly been associated with inbreeding and high levels of homozygosity ([19]).
The aims of this study were to: (i) evaluate the selfing rate and impact of inbreeding depression in E. globulus and to what extent family genetic background negatively impacted growth in self-crossed progenies; (ii) study the prevalence of outside pollen contamination in progenies collected in a seed orchard; and (iii) assess whether morphological abnormalities detected in some of the offspring were related to their level of inbreeding.
To accomplish this, several field tested open-pollinated family progenies between 2 and 5 years were evaluated for growth rate and presence of abnormalities. Given the relatively young age, height, rather than diameter, was considered a better proxy of later age tree volume. Early height was shown to be strongly correlated with later age volume growth, with correlations ranging between 0.8 and 0.9 ([5]). These families were collected in an E. globulus seed orchard and their full pedigree was reconstructed based on molecular markers. An individual tree mixed linear model accounting for all pedigree information was fitted to quantify the importance of inbreeding depression and how much it varies between families.
Materials and methods
Seed orchard
The tested E. globulus progeny was collected from a clonal seed orchard located at Herdade de Espirra, near the city of Pegões in southern Portugal (38° 41′ N, 08° 12′ W). The orchard occupies 1.7 ha and was established in 2008. It is located on a flat area, with an initial spacing of 5 × 5 m, and is surrounded by native stone pine (Pinus pinea) and cork oak (Quercus suber) stands, with the nearest E. globulus trees located at least 600 m away. The orchard consists of 27 E. globulus parent trees, with variable number of copies (ramets) per genotype, ranging from 1 to 60. These are first- and second-generation selected material from RAIZ eucalypt breeding program.
Families and field trials
Up to eleven open pollinated families were collected in the orchard, at three consecutive years (2014, 2015 and 2016). These families were selected because either they resulted from the most represented mothers in the orchard, with between 14 and 40 flowering ramets each (see Tab. S1 in Supplementary material for detailed information) or they had progeny with large number of abnormal phenotypes (see below).
The progeny from these families (n=324) was tested across five locations, covering a range of sites in Central and Southern Portugal. All trials were designed as randomized complete blocks with up to 9 replicates, with each family represented in each replicate by 1 to 5 tree plots, with a standard spacing of 0 × 2 m (see Tab. S2 in Supplementary material for trials’ edaphoclimatic conditions). As a result of initial variation in progeny numbers and subsequent mortality, each family was represented in the dataset varying between 1 and 24 offsprings per site.
Phenotypic measurements
Survival across all trials was high being on average 95%. The mixed model analysis of growth included all trees with normal phenotype coming from the 7 most representative families of the orchard. They were measured for height, with ages between 1.6 to 4.5 years (n=283 - Tab. 1). Since the percentage of trees in the field trials presented abnormalities were initially low, to increase sample size, four extra families were added to the study (but not included in the mixed model - see below). Normal phenotypes were ascribed to individuals having the typical juvenile eucalypt canopy, with opposite juvenile leaves, lanceolate and symmetrical and with a single axillary bud at the base of each leaf. In contrast, abnormal phenotypes were ascribed to trees having an excessive proliferation of axillary buds and a loss of apical dominance, resulting in a mallee or bushy appearance.
Tab. 1 - Details of the trial location, age of measurements and experimental design in the five field trials included in the mixed model analysis. Number of Randomized complete blocks and families tested, and the total across family progeny measured are also given (n=283).
| Trial Location | Seed year |
Age | # Replicates | # Families | Total progeny size |
|---|---|---|---|---|---|
| Fundão | 2014 | 4.5 | 4 | 4 | 53 |
| Abrantes | 2015 | 3.2 | 9 | 4 | 75 |
| Oleiros | 2015 | 3.2 | 9 | 5 | 50 |
| Gorda | 2016 | 2.3 | 9 | 6 | 54 |
| Nisa | 2016 | 1.6 | 9 | 6 | 51 |
Molecular analysis
DNA was extracted from leaves of 324 progeny and 27 putative parents in this study, with an optimized CTAB protocol (as described in [22]). Spectrophotometry was used to calculate DNA concentrations and ensure dilutions equal to an optimal concentration of 10 ng µL-1. A total of 12 microsatellite (SSR) markers were selected, covering all linkage groups in the Eucalyptus sp. genome ([45]), and were amplified in four multiplex sets (see Tab. S3 in Supplementary material). The PCR products detection was established by electrophoresis with use of ABI 3130 Genetic Analyzer® (Applied Biosystems, Foster City, CA, USA) followed by alleles length detection using the Peak Scanner software v. 2.0.
Parental assignment (i.e., pedigree reconstruction) was carried out based on the allele data for all the progeny belonging to the seven open pollinated families and considering all putative parental individuals in the orchard. The assignment was conducted using standard maximum likelihood methods implemented in CERVUS® 3.4 ([35], [29]). The analysis determined the critical value of Delta (D) for each confidence in the paternity and maternity analysis, defined as the difference between the “LOD score” of the first most likely parent candidate and the “LOD score” of the second most likely candidate. Significance of the paternity tests simulated by CERVUS software was based on a confidence level of 80% (relaxed) to 95% (strict), as suggested by Marshall et al. ([35]), using 50.000 repetitions, 0.01 as the ratio of genotyping errors, and 90% as the proportion of pollen donors sampled within each population (given the high degree of isolation of the orchard). The minimum number of loci necessary to determine the paternity of a seed was set at six.
If a seedling had no assigned pollen donor from any of the orchard parental population, pollen parents were assumed to be originated from unrelated trees from outside the orchard, hence resulting in foreign pollen contamination. In pedigree reconstruction for each seed, the possibility of having one single mismatch (or reading error) was considered in the simulation, as recommended in Kalinowski et al. ([29]).
Data and statistical analyses
Selfing and contamination rates were estimated for the most representative families in each of the three different seed collection years. Percentage selfing per open-pollinated family (i.e., per maternal parent) was estimated by dividing the number of selfed offspring, by the total number of offspring in that family. Similarly, contamination rates were calculated by dividing the number of offspring sired by foreign pollen by the total number of offspring in the family. As mentioned, pollen was considered foreign if it did not match any of the genotypes present in the orchard. The inbreeding coefficient of each offspring (F) was estimated based on the complete reconstructed pedigree. The package “optiSel” in R ([49]) was used to calculate the expected F values for each tree. Group co-ancestry (Θ) for parents, progeny and Status Number were calculated as described by Cockerham ([12]), Lindgren et al. ([32]) and Lindgren & Mullin ([33]). Coancestry or coefficient of kinship is the probability that a gene taken at random from an individual will be identical by descent with a random homologous gene from another individual, including identical individuals (self-coancestry). Group coancestry (θ), as used here is the average of all possible pairwise coancestries, including self-coancestry and reciprocals between all trees in the pedigree set. By definition, the status number (Ns) is half the inverse of the group coancestry, thus Ns = 0.5/Θ. Only when all seed orchard genotypes contribute equally, in addition to being unrelated and noninbred, will Ns equal the census number in the orchard.
Since trials at the time of measurement had different ages, means and variances of height varied considerably between trials. Mean annual height increments, rather than total height, was used in the linear model, to ensure measurements are more comparable and variances are homogeneous across sites. The following mixed model was fitted (eqn. 1):
where y is the vector of height increments, f the vector of fixed effects, which includes the effects due to site, replicate within site, and population mean inbreeding depression slope (bF), fitted as a covariate; a is the vector of tree’s additive genetic effect, with a ~ NID (0, Aσ2a), where A is the average numerator relationship matrix (a matrix with the additive genetic relationships between all trees in the pedigree) and σ2a is the additive genetic variance. The bFi is the vector of genetic inbreeding depression slopes, fitted as a random covariate for each genotype, with bFi ~ NID (0, Iσ2bFi) where σ2bFi is the inbreeding depression slope variance and e the vector of residual effects with e ~ NID (0, Iσ2e), where σ2e is the residual error variance. The X and Zi are incidence matrices relating observations to the respective effects. To test whether the assumption of homogeneous residual variances could be held across the five sites, the above model was compared with an alternative model with a block diagonal R structure (with one residual section for each site) and fitted as R = ï 5j=1 Rj where Rj= Iσ2e. We tested the null hypothesis that error variances are equal among sites, using a likelihood ratio test. The LRT=4.876 (with 4 df; p=11%), hence we accepted the assumption of homogeneous variances across sites. Further significance of random effects (a and bFi), was also tested based on a Likelihood Ratio Tests, assuming 0.5 df as suggested by Self & Liang ([46]) Significance of fixed effects was tested based on a scaled Wald F statistic, following the Kenward & Roger ([30]) simplification, as implemented in AsReml v. 4.2 ([20]).
This model assumed mean annual height increments from height measurements taken at different ages (as height measurements divided by their corresponding age) would be an expression of the same trait. High genetic correlations between height at different early ages have been reported for E. globulus ([5]).
Across-site heritability for height growth was estimated without explicitly fitting a genotype by site interaction term, since the number of genotypes was small and there was unbalance in their representation across sites. It is expected that such effects, if present, would be captured in the residual term. Heritability was estimated as (eqn. 2):
Taylor series expansion was used to obtain approximate standard errors for the variance parameters estimated from the linear mixed model, as well as for derived linear combinations or ratios of the variance estimates, as implemented on the “vpredict” function in AsReml v. 4.2 ([34], [20]).
The association between frequency of abnormal phenotypes and inbreeding was tested based on a χ2 independence test, using the R function “chisq.test” with data from Tab. S4 (Supplementary material) as input.
Results and Discussion
Allele counts and heterozygosity
Amplification was successful across the 12 SSR markers, corroborating their importance in paternity tests in E. globulus reported previously ([16]) and in kinship analyses ([45]). Overall, only 1.6% of the individuals had two missing SSR amplifications, 3.7% had three missing microsatellites, 1.2% had four and 0.3% had six. Allele diversity was high, with an average of 15.5 alleles per loci across all SSRs and with most showing eight or more alleles (Tab. 2). Such allelic diversity is somewhat higher than that found for the native land race in Portugal ([18]) and closer to the values reported across the species natural distribution ([16]). This is expected since parents represented in the orchard are derived from various native provenance backgrounds. Heterozygosity was also high with He = 0.86 (Tab. 2), and similar to levels reported in domesticated and wild populations of E. globulus ([16]).
Tab. 2 - Details of the nuclear SSR markers used in the analysis, and the number of alleles, the percentage of null alleles and the expected heterozygosity (He) found across the seven open-pollinated families tested.
| Microsatellite | No Alleles |
Null (%) |
He |
|---|---|---|---|
| Embra11 | 17 | 0.10 | 0.87 |
| Embra119 | 14 | 0.14 | 0.86 |
| Emcrc8 | 17 | 0.04 | 0.88 |
| Emcrc7 | 8 | 0.08 | 0.77 |
| Embra23 | 15 | 0.08 | 0.90 |
| Embra41 | 15 | 0.07 | 0.86 |
| Embra227 | 10 | 0.04 | 0.78 |
| En15 | 11 | 0.03 | 0.84 |
| Es76 | 18 | 0.07 | 0.90 |
| Eg65 | 18 | 0.05 | 0.89 |
| Embra37 | 23 | 0.06 | 0.91 |
| En12 | 17 | 0.08 | 0.87 |
| Average | 15.25 | 0.07 | 0.86 |
Seed orchard characterization
In the present study, at the time of measurement, between 2 and 4 years, the degree of selfing across families was on average 30%. This estimate is in the upper range of what has been reported in other orchards ([38], [21]). Selfing rates varied considerably between collection years (Tab. 3). In 2014, no selfing was reported. This is a surprising result since it was a year of particularly poor flowering (data not shown), where high rates of selfing would be expected. In the following years, selfing was considerably higher. Since survival in the trial was consistently high at all sites (greater than 95%), these differences are unlikely to have been caused by selective mortality of some of the selfs. Selfing percentages also varied among different parents, but again the pattern was unclear. In the 2015 seed year, selfing rates were low (at around 10%) for three of the families (M74, M07 and M13), and very high (around 90%) for two other families (M49 and M28). In the following seed year (2016), family M28 showed a relatively low selfing rate (14.3%), much lower than in the previous year. These results suggest other factors such as flower synchrony and the activity of pollinators at the time of stigma receptivity may play a significant role.
Tab. 3 - Total number of progenies sampled from each family (ID Mother) and seed collection year, as well as the number and respective percentage of selfed and contamination with foreign pollen. The trial’s location where the progenies were tested are also given (in parentheses). The progeny listed only includes the most representative families within each year (n = 311).
| Year (Trial) | ID Mother | Progeny | Self | % Selfing |
Contaminated | % Contamination |
|---|---|---|---|---|---|---|
| 2014 (Fundão) | M74 | 9 | 0 | 0.0 | 0 | 0.0 |
| M07 | 13 | 0 | 0.0 | 0 | 0.0 | |
| M13 | 15 | 0 | 0.0 | 0 | 0.0 | |
| M18 | 18 | 0 | 0.0 | 0 | 0.0 | |
| 2015 (Abrantes and Oleiros) | M74 | 27 | 3 | 11.1 | 0 | 0.0 |
| M07 | 31 | 3 | 9.7 | 5 | 16.1 | |
| M13 | 34 | 3 | 8.8 | 5 | 14.7 | |
| M49 | 27 | 26 | 96.3 | 1 | 3.7 | |
| M28 | 17 | 15 | 88.2 | 1 | 5.9 | |
| 2016 (Gorda and Nisa) | M74 | 19 | 7 | 36.8 | 3 | 15.7 |
| M07 | 15 | 1 | 6.7 | 5 | 33.3 | |
| M13 | 20 | 2 | 10.0 | 4 | 20 | |
| M89 | 24 | 16 | 66.7 | 0 | 0.0 | |
| M49 | 21 | 12 | 57.1 | 2 | 9.5 | |
| M28 | 21 | 3 | 14.3 | 4 | 19 |
Contamination rates, from pollen outside the orchard, were low in absolute terms (average of 10% across years) but seem to be increasing over time. It was 0% in 2014, 8% in 2015 and 16% in 2016 (Tab. 3). Since Eucalyptus is mostly an insect pollinated species, distance of pollen dispersion is more limited than for wind pollinated species and dependent of the involved specific insect populations with maximum recorded pollen dispersal distances of a few hundred meters only ([4], [21], [17]). Considering the distance between the orchard and any other putative E. globulus tree in the neighborhood in the present study was over 600 m, the contamination rates found are higher than expected. This highlights the importance to better understand the pollination ecology and in particular the relevance of long-distance pollen dispersal mechanisms in eucalypt seed orchards ([7], [27]).
The causes for the substantial annual variation in selfing and to a lesser extent in contamination rates are hard to interpret since no data on the phenology of trees inside and around the orchard, and insect activity has been recorded in detail. The flowering synchrony, the length and intensity of flowering, the weather conditions at critical pollination periods and the intensity of insect activity are all plausible factors affecting pollen dispersal in a seed orchard, hence influencing outcrossing and contamination rates.
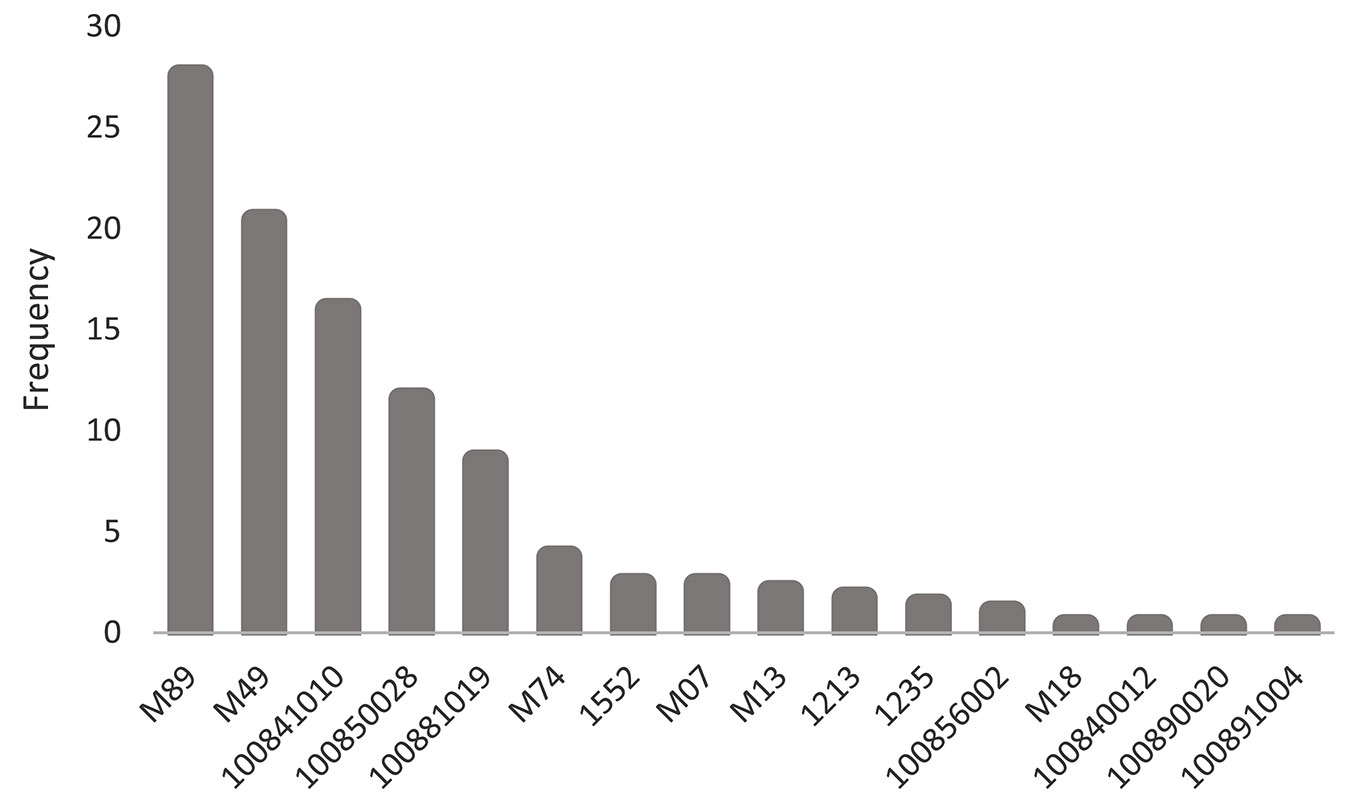
All unrelated base parents in the orchard were assumed to have an inbreeding of F=0. However, some parents in the orchard are second- and third-generation selections, with some degree of kinship among them although not inbred themselves. The average co-ancestry among the 15 orchard parents contributing to the studied progeny, derived from pedigree records, was greater than zero (θP=0.05) and corresponds to a Status Number of NS=9.28, less than the census number of N=15. Their progeny (N=283), on the other hand, included crosses between some of these related parents, as well as from self-pollination. Therefore, its group co-ancestry was higher (θOff=0.075) and the expected inbreeding was greater than zero (FOff=0.17), although the corresponding Status Number remained similar (NOff=9.27). This build-up of inbreeding in the orchard’s progeny was mostly the result of selfing, since very few progenies have been the result of outcrosses between related parents. There was also a substantial unbalance in the frequency of full- and half-sibs among families and in the relative pollen contribution to the progeny by the putative paternal parents (Fig. 1), with the top three paternal parents contributing to 65% of the progeny. Given the crossing scheme present in the dataset, the observed intra-class correlation amongst open-pollinated (OP) sibs was r=0.51, ranging across families between 0.32 and 0.83 (Tab. S5 in Supplementary material). In other words, open-pollinated sibs were on average as related as full-sibs (where r=0.5), a clear departure from the often-assumed half-sib kinship in open pollinated progeny studies.
Fig. 1 - Frequency of male contribution (pollen) to the progeny in the field-tested offspring. The counting includes progeny from seed collected over a period of three flowering seasons (2014-2016) and tested in five different locations, with ages ranging between 1.6 and 4.5 years.
Inbreeding depression for height growth
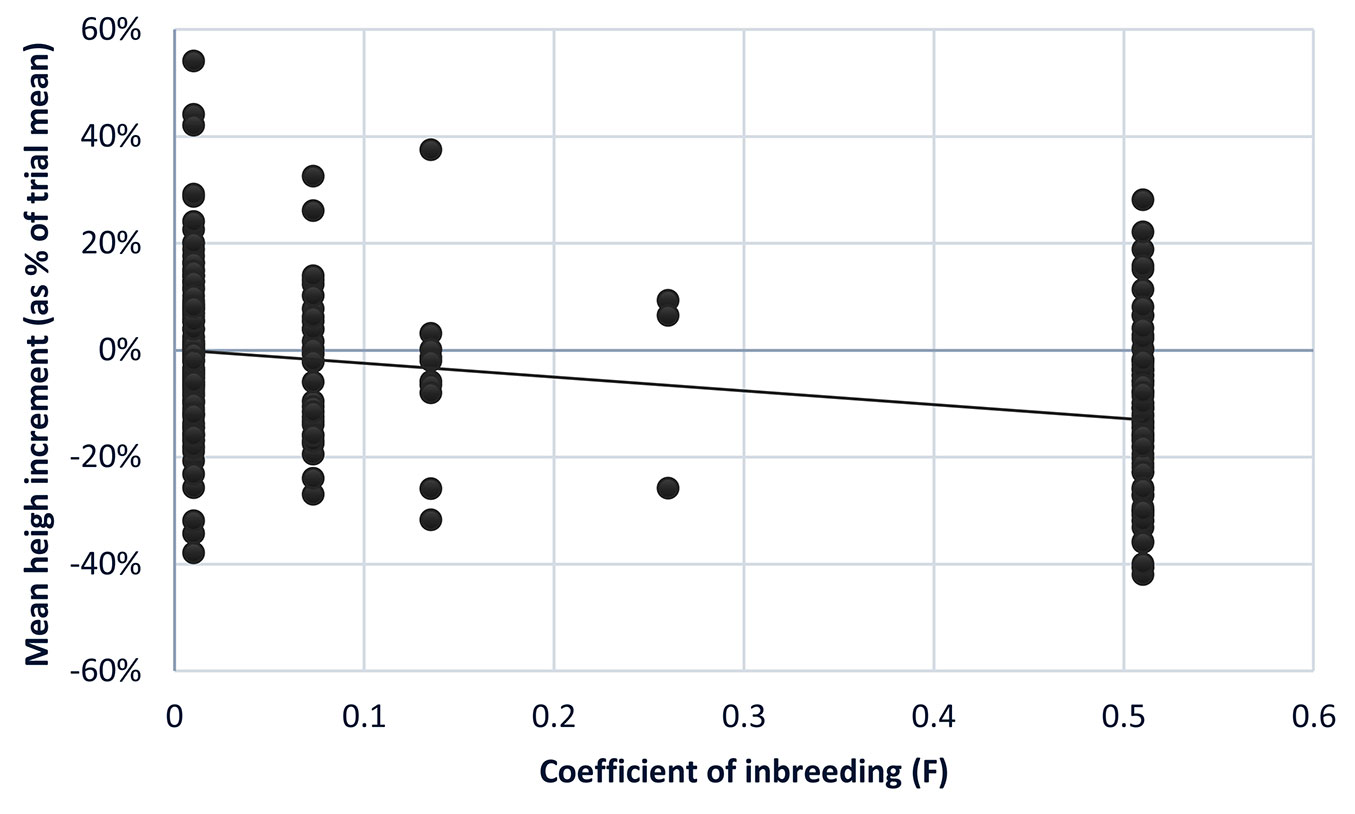
The reconstructed pedigree for all sampled trees allowed the estimation of the inbreeding coefficient (F) for each tree. Of the 283 trees with normal phenotypes used in the mixed model analysis, 54% had unrelated parents (F=0), 11% had F=0.0625, 4% had F=0.125, 1% had F=0.25 and 30% had F=0.5. Mean annual increment across the five trials was 2.9 m, with site effects and replicate effects within site being significant (p<0.001). The population mean inbreeding depression slope (bF) was significant (bF = -0.867; p = 0.017). In other words, for F=1, mean annual height growth is expected to be reduced on average by 0.87 m ± 0.32. Progeny resulting from self-pollination, which has an F=0.5, would be expected on average to be 0.43 m year-1 smaller in height when compared with outcrossed progenies from unrelated parents (Fig. 2), which corresponds to an inbreeding depression of 14.7%. These are similar to estimates published previously for growth in E. globulus ([24], [15], [39]). However, since the present study is based on young trees, between 1.6 and 4.5 years old, is likely to be an underestimation, as the impact of inbreeding depression is likely to increase with time due to increasing inter-tree competition ([47], [48], [36], [15], [50]). This highlights the importance of ensuring self-pollination rates are kept to a minimum in seed orchards.
Fig. 2 - Mean annual height increments (given as percentage of the trial means) for all trees in the five-progeny trial (n=283), relative to their corresponding inbreeding coefficient.
The inbreeding depression slopes for each of the seven maternal parents studied (bFi) showed a consistent decrease in growth with inbreeding (i.e., they all have a negative coefficient), with its magnitude varying from bFi = -0.4 to -1.4 (Tab. 4). For example, progeny from parent M74 was substantially more impacted by inbreeding, with an estimate annual height loss of -1.4 m when F=1 (an inbreeding depression in selfs of 24%), whereas M13 was more tolerant to inbreeding with bF=-0.43 (or 7% depression in selfs - Tab. 5).
Tab. 4 - Variance components for the random effects in the mixed linear model for annual height growth and corresponding likelihood ratio tests (LRT) and probability of significance. Effects include the trees’ additive genetic effects (a), the deviation of inbreeding depression slopes per genotype, relative to the population mean slope (bFi), and the residual effects (e).
| Effects | Variance | LRT | Prob |
|---|---|---|---|
| a | 0.017 | 7.12 | 0.008 |
| b Fi | 0.476 | 6.16 | 0.013 |
| e | 0.064 | - | - |
Tab. 5 - Estimates of the solutions for the overall and site mean, the parent’s additive genetics effects (a), the mean population inbreeding coefficient slope (bF), and the individual parent inbreeding coefficient slopes (bFi) for the seven parents in the study.
| Effects | Mother ID | Estimate | Standard error |
|---|---|---|---|
| Mean | - | 2.929 | 0.145 |
| Site | 394 | 2.929 | 0.145 |
| 407 | 2.367 | 0.156 | |
| 416 | 3.355 | 0.210 | |
| 421 | 2.853 | 0.185 | |
| 423 | 1.596 | 0.193 | |
| Parents breeding value (a) | M74 | 0.231 | 0.089 |
| M18 | 0.032 | 0.106 | |
| M49 | 0.097 | 0.099 | |
| M28 | 0.013 | 0.099 | |
| M07 | -0.117 | 0.087 | |
| M13 | -0.024 | 0.086 | |
| M89 | -0.012 | 0.080 | |
| bF | -0.867 | 0.327 | |
| b Fi | M74 | -1.404 | 0.392 |
| M18 | -1.310 | 0.628 | |
| M49 | -0.795 | 0.375 | |
| M28 | -0.580 | 0.392 | |
| M07 | -0.743 | 0.460 | |
| M13 | -0.432 | 0.423 | |
| M89 | -0.724 | 0.386 |
Despite this large variation in the inbreeding depression slope between parents, an effect fitted as a random term in the model, differences between them and the mean population slope bF = -0.867 were only moderate (LRT = 6.16; p=0.013; df = 0.5; and variance of σbFi2 = 0.476). Although indicative of a genetic effect underlying the inbreeding depression slopes, the statistical power of the test was surely affected by the relatively low number of offsprings per parent and per class of inbreeding. Further evidence of a genetic effect on inbreeding depression should be confirmed in future studies.
Across-site additive genetic effects for height were significant, with a variance of σA2 = 0.017 (LRT=7.12; p=0.008 - Tab. 4), which corresponds to an estimated heritability for height of h2=0.21 (standard error=0.16), an estimate well within the reported for the species and trait ([5], [24], [41], [14]). Given the very limited sample size and unbalance in the crossing scheme, the linear model fitted only accounted for additive genetic effects and did not include dominance or other non-additive genetic effects. Since dominance effects are known to be of importance for growth traits in eucalypts ([13], [2]), and if unaccounted, likely to cause an upward bias in the heritabilities ([5]), these estimates are expected to be somewhat over-estimated.
Abnormalities
Among the 324 trees assessed, 30 were classified as having abnormal phenotypes, showing dwarfism and a mallee appearance, as a result of partial or total loss of apical dominance and excessive proliferation of axillary buds. However, it was clear from the pedigree records that there was no obvious association between such abnormalities and selfed individuals, as many abnormal phenotypes were found in unrelated outcrossed progeny (where F=0). The chi-square test confirmed the independency observed between abnormal phenotypes and estimated levels of inbreeding (null hypothesis: χ2[4] = 2.4277; p = 0.6576). To clarify any putative association between abnormal phenotypes and parents, we looked at crosses with 10 or more offsprings. This analysis showed however that abnormal phenotypes were more frequent in specific, but otherwise unrelated, crosses (Tab. 6). For example, cross between M49 and M28 produced seven abnormal phenotypes out of a total of 22 trees (an incidence of 32%), whereas selfs from the same mother tree (M49) only had one abnormal phenotype out of 38 trees present in the trials. Also, cross between parent trees 100841010 and M18, although with a lower progeny size, showed four abnormal trees out of a total of 10. Genetically, this could be a consequence of particular deleterious effects between specific alleles present in the two parents. If these alleles are rare, this would be highly unlikely to occur across several independent crosses, as found in our study. This question regarding abnormal phenotypes deserves further clarification with an experiment that includes a larger sampling size.
Tab. 6 - Progeny with normal or abnormal phenotypes resulting from crosses with 10 or more offspring present in the five progeny trials (aged between 1.6 and 4.5 years). Parent A and Parent B columns are not indicative of female or male parents and merely represent the cross between the two clones.
| Parent A | Parent B | Normal phenotype | Abnormal phenotype |
|---|---|---|---|
| M49 | M28 | 15 | 7 |
| M49 | M49 | 37 | 1 |
| 100841010 | M18 | 6 | 4 |
| M07 | M89 | 12 | 0 |
| M07 | 100841010 | 11 | 1 |
| M07 | 100881019 | 13 | 0 |
| M13 | M89 | 33 | 1 |
| M89 | M89 | 13 | 3 |
| M28 | M28 | 18 | 0 |
| M74 | M89 | 16 | 0 |
| M74 | 100841010 | 10 | 0 |
| M74 | M74 | 10 | 0 |
Conclusion
The studied open-pollinated E. globulus seed orchard progeny presented a substantial degree of self-pollination, accounting for an average of 30% in the 2- to 4-year-old progeny from seed crop collected over three years. Such selfing resulted in an average inbreeding depression of 14.7% in height growth. However, the magnitude of inbreeding depression differed amongst genotypes, ranging between 7% and 24% when comparing selfed with outcrossed progeny from unrelated parents. Preferential harvesting of seed from maternal parents less affected by inbreeding depression should therefore be a useful measure to improve seed quality from orchards. Another important result was that no relation has been found between the presence of anomalous phenotypes and selfing. Instead, a high frequency of abnormalities has been found in specific but unrelated crosses. These finding highlight the importance of understanding the reproductive biology, the genetic basis of reproductive success, the gene flow in orchards and the merits of using genetic markers to regularly monitor factors such as rates of contamination, selfing and inbreeding depression.
Acknowledgements
All authors contributed to the conceptualization, methodology definition, data analysis and interpretation, as well as the review and edition of the manuscript. All authors have read and agreed to the published version of the manuscript. Laboratory work was performed by JF supervised by HT, fieldwork supervised by JA; writing of the original draft was first outlined by JF and HT, review and editing was performed by all the authors.
The authors would like to thank to The Navigator Company for providing access to the study sites, to Luis Ferreira and José Cardoso for trial assessments and Fátima Cunha for expert lab assistance.
This research was funded by Fundação para a Ciência e a Tecnologia (FCT, Portugal), FCT/MCTES, through the financial support to CESAM (UIDP/50017/2020, UIDB/50017/2020 and LA/P/0094/2020) and the financial support to cE3c (UIDB/00329/20 20).
Conflicts of Interest
The authors declare no conflicts of interest.
References
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
CrossRef | Gscholar
Authors’ Info
Authors’ Affiliation
Helena Trindade 0000-0002-1209-2622
Universidade de Lisboa, Faculdade de Ciências, Departamento de Biologia Vegetal, 1749-016 Lisboa (Portugal)
José Araújo
Nuno Borralho 0000-0001-5606-7878
Cristina Marques 0000-0003-4630-8652
Raiz, Forest and Paper Research Institute, Quinta de S. Francisco, Apartado 15, 3801-501 Eixo (Portugal)
cE3c - Center for Ecology, Evolution and Environmental Change; CHANGE - Global Change and Sustainability Institute - Formerly CESAM, Centro de Estudos do Ambiente e do Mar, Faculdade de Ciências, Lisbon (Portugal)
Corresponding author
Paper Info
Citation
Faia J, Costa J, Araújo J, Borralho N, Marques C, Trindade H (2022). Impact of inbreeding on growth and development of young open-pollinated progeny of Eucalyptus globulus. iForest 15: 356-362. - doi: 10.3832/ifor4012-015
Academic Editor
Alberto Santini
Paper history
Received: Nov 05, 2021
Accepted: Jul 06, 2022
First online: Sep 20, 2022
Publication Date: Oct 31, 2022
Publication Time: 2.53 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2022
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 27920
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 23743
Abstract Page Views: 2107
PDF Downloads: 1592
Citation/Reference Downloads: 4
XML Downloads: 474
Web Metrics
Days since publication: 1267
Overall contacts: 27920
Avg. contacts per week: 154.25
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
(No citations were found up to date. Please come back later)
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Pollen contamination and mating patterns in a Prosopis alba clonal orchard: impact on seed orchards establishment
vol. 12, pp. 330-337 (online: 14 June 2019)
Review Papers
Genetic diversity and forest reproductive material - from seed source selection to planting
vol. 9, pp. 801-812 (online: 13 June 2016)
Research Articles
Genetic diversity of core vs. peripheral Norway spruce native populations at a local scale in Slovenia
vol. 11, pp. 104-110 (online: 31 January 2018)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Clonal structure and high genetic diversity at peripheral populations of Sorbus torminalis (L.) Crantz.
vol. 9, pp. 892-900 (online: 29 May 2016)
Research Articles
Growth, spring phenology and stem quality of four broadleaved species assessed in provenance trials in the Netherlands - Implications for seed sourcing
vol. 18, pp. 242-251 (online: 22 September 2025)
Research Articles
Delineation of seed collection zones based on environmental and genetic characteristics for Quercus suber L. in Sardinia, Italy
vol. 11, pp. 651-659 (online: 04 October 2018)
Research Articles
Age trends in genetic parameters for growth and quality traits in Abies alba
vol. 9, pp. 954-959 (online: 07 July 2016)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword