Comparison of genetic parameters between optimal and marginal populations of oriental sweet gum on adaptive traits
iForest - Biogeosciences and Forestry, Volume 11, Issue 4, Pages 510-516 (2018)
doi: https://doi.org/10.3832/ifor2450-011
Published: Jul 18, 2018 - Copyright © 2018 SISEF
Research Articles
Collection/Special Issue: COST Action FP1202
Strengthening conservation: a key issue for adaptation of marginal/peripheral populations of forest trees to climate change in Europe (MaP-FGR)
Guest Editors: Fulvio Ducci, Kevin Donnelly
Abstract
Genetic parameters of 9 oriental sweetgum (Liquidambar orientalis Mill.) populations were estimated at a common garden test. Open-pollinated seeds were collected from 16-27 families for each population. The common garden test was established in 2009 using a randomized complete block design in 25 blocks with single tree plot, with each block included 223 families. Breast height diameter, height and crown diameter were measured at the age of five. The purpose of study was to compare the genetic parameters of optimal and marginal populations and to assess the findings for genetic conservation. The study revealed significant variability in all traits evaluated. In variance components, variation among populations was three times higher than that of families. Individual heritability estimates for breast height diameter, height and crown diameter pooled across the whole dataset (marginal and optimal populations) were 0.21 ± 0.04, 0.27 ± 0.04 and 0.11 ± 0.03 and additive genetic coefficients of variation were 13.4%, 9.1% and 7.1%, respectively. Individual heritability estimates for breast height diameter, height and crown diameter in marginal and optimal populations were 0.27 ± 0.10, 0.19 ± 0.08 and 019 ± 0.08 and 0.19 ± 0.04, 0.29 ± 0.05 and 0.09 ± 0.03, respectively. Additive genetic coefficients of variation for breast height diameter, height and crown diameter were 16.7%, 8.3% and 10.8% in marginal and 12.8%, 9.1% and 6.2% in optimal populations, respectively. While breast height diameter and crown diameter were more heritable for marginal populations, height was more heritable for optimal populations. These findings are discussed in terms of genetic conservation of oriental sweet gum.
Keywords
Liquidambar orientalis, Genetic Variation, Individual Heritability, Gene Conservation, Climate Change
Introduction
Oriental sweet gum (Liquidambar orientalis Mill.) is a relic and endemic species of Turkey. The natural distribution area of the species is only 3.200 hectares between 0-1000 meters in elevation in southwest Turkey. Adult trees with breast height diameter more than 20 cm are usually wounded repeatedly to produce valuable balsam ([34]). Since almost all trees have been continuously injured to produce balsam, fact that may be result to their dead, the species is at the risk of extinction. Therefore, it was identified as a priority species for conservation and a technical guideline for its conservation and use was prepared by the European Forest Genetic Resource Programme (EUFORGEN - [2]).
Oriental sweet gum is an economically important species, due to the natural balsam producing ability which is exceptional among forest trees. Sweet gum oil is used in pharmaceutical, chemical, and cosmetic sectors. It is a raw material for in the production of cinnamyl alcohol and acid and it is used as a stabilizer for soap and cosmetic products. Almost all of the produced oil is exported ([2]). Due to over exploitation, oil production has dramatically declined from 180 tons in 1950 to 1,113 kg in 2013 ([36], [24]).
The natural distribution area of oriental sweet gum comprises of areas with very productive deep soil and of riparian lowland habitats; areas that attract interest for farming. For this reason, a great portion of the oriental sweet gum lands has been illegally transformed in agricultural fields. Due to habitat loss by deforestation and drainage of lowland of the Aegean Region, the natural distribution area of the oriental sweet gum has been decreased from 7,000 ha to 3,200 ha since 1947 ([2]).
Knowledge on the existing genetic variation within and between populations is essential for the species management, its domestication and the application of breeding and conservation strategies ([7], [23]). The molecular markers and the common garden tests are two powerful tools applied for the characterization of genetic variation in forest tree populations. While genetic markers are beneficial to convey genetic drift, gene migration and mating system, common garden tests are the ones providing information for quantitative traits and more explanatory for defining adaptive variations ([12], [38]).
Recently, a comprehensive genetic diversity research project has been conducted with oriental sweet gum by using molecular markers. For this purpose, 18 populations were studied covering the whole distribution area of oriental sweet gum ([36]). The results revealed that the difference among populations was high and the gene flow among populations was low. Such a result was not surprising, because the populations were often separated or disconnected. Velioglu et al. ([36]), recommended that 9 out of the 18 populations should be conserved as ex situ genetic resources. Then, a common garden test was established in 2009 with those 9 populations suggested for ex situ conservation. Many researches using molecular markers have already been carried on ([25], [26], [3], [19], [27]). Except for Albayrak & Arican ([3]), researchers studied the whole 18 populations using molecular markers, finding divergences and similarities of populations, and recommending some populations for in situ or ex situ genetic conservation. Albayrak & Arican ([3]) found also similarities and divergences among five populations. Despite such many molecular investigations, there has been no studies on quantitative traits of oriental sweet gum. Indeed, the best way to test for genetic differences among populations within a species is to conduct common garden experiments ([29]). Information from common garden test may improve our understanding of local adaptations to environmental factors, as well as our predictions regarding the impact of climate change on tree fitness and community composition ([37]).
Species are expected to have highest abundance at the core areas of their distribution, with smaller and more disjunct populations found at the margins of distribution ([8]). Understanding the genetic characteristics of plant populations at the margins of a species’ natural range is important because the peripheral populations are likely to preserve their adaptive and evolutionary potential under the anticipated global climate change conditions ([22], [1], [11], [28]). Whether geographically central populations differ in their genetic diversity and differentiation levels from their marginal counterparts remains an open question. However, reviews of empirical studies have reported contradictory results on the patterns of distribution of genetic variation in marginal vs. central populations ([8], [22], [15], [35]). Eckert et al. ([11]) reviewed 134 studies, representing 115 species, that used molecular and biochemical genetic markers, and found that in most cases, the difference in genetic diversity between central and peripheral population was not large. Molecular studies that related optimal and marginal population were abundant as mentioned above, though studies based on quantitative traits were limited ([31], [21]) and not directly focused on differences between optimal and marginal populations ([30]). Oriental sweet gum is a vulnerable species, therefore, in addition to molecular studies, information on quantitative traits of marginal and optimal population is fundamental for its short and long term genetic conservation.
In this study, 5-year-old seedlings from 9 different populations of Liquidambar orientalis were measured in a common garden experiment and evaluated with the aim of: (a) comparing some genetic parameters between marginal and optimal populations; (b) assessing the implications for conservation of oriental sweet gum.
Materials and methods
Genetic material experimental design
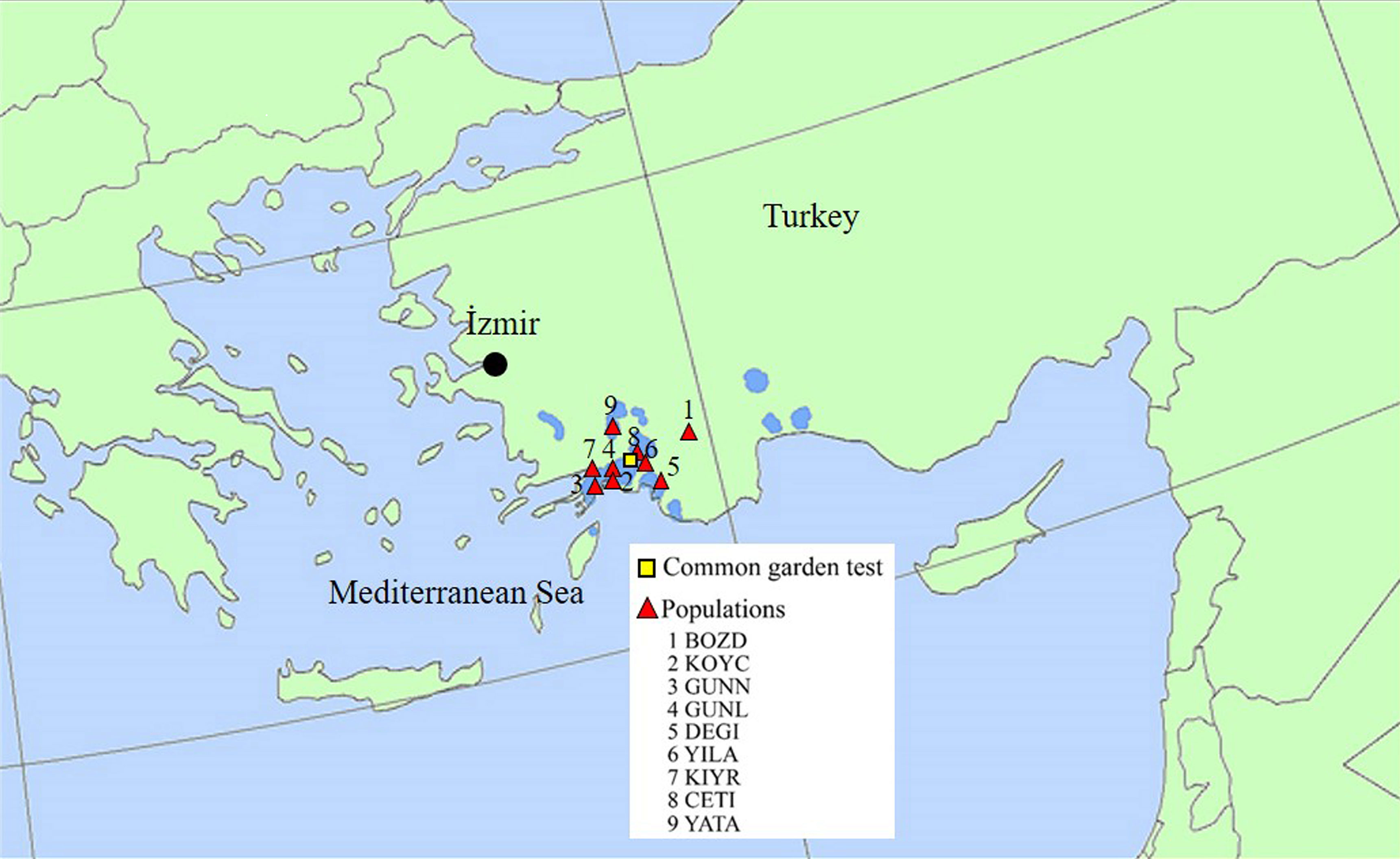
Nine populations were used in the common garden test, as recommended by Velioglu et al. ([36]). BOZD population is located at the highest altitude, while YATA is the northernmost population; other populations are mostly situated at the center of the species’ distribution area (Fig. 1).
Fig. 1 - Location of the nine populations of Oriental sweet gum analyzed and of the common garden test. Base map from EUFORGEN ([13]).
Wind-pollinated seeds were previously collected from mother trees from natural populations, with a minimum horizontal distance of 100 m between sampled trees in each population and a maximum elevation difference of 300 m. Sampled trees were either located in stands, where oriental sweet gum is the dominant species, or scattered along the creeks in riparian stands, where the above minimum elevation difference criterion was sometimes disregarded due to the narrow elevation zone. The mean altitude of the sampled sites ranged among 5 and 1,100 m a.s.l. (Tab. 1).
Tab. 1 - Geographical data of the studied populations.
| Population origin | Code | No. of families |
No. of seedlings |
Population type |
Altitude (m a.s.l.) |
Lat N Long E |
|---|---|---|---|---|---|---|
| Acipayam-Bozdag | BOZD | 16 | 349 | Stand | 1100 | 37° 18′ 58″ 29° 17′ 31″ |
| Koycegiz- Koycegiz | KOYC | 26 | 574 | Stand | 10 | 36° 57′ 30″ 28° 42′ 09″ |
| Marmaris-Gunnucek | GUNN | 24 | 499 | Stand | 5 | 36° 51′ 30″ 28° 17′ 38″ |
| Fethiye-Gunlukbasi | GUNL | 27 | 601 | Stand | 5 | 36° 40′ 12″ 29° 07′ 37″ |
| Marmaris-Degirmenyani | DEGI | 25 | 578 | Stand | 5 | 36° 49′ 21″ 28° 08′ 19″ |
| Mugla-Yilanli | YILA | 25 | 548 | Along creek | 250 | 37° 02′ 27″ 28° 10′ 21″ |
| Mugla-Kiyra | KIYR | 25 | 556 | Along creek | 50 | 37° 03′ 42″ 28° 32′ 47″ |
| Marmaris-Cetibeli | CETI | 26 | 582 | Along creek | 30 | 36° 58′ 57″ 28° 17′ 38″ |
| Mugla-Yatagan | YATA | 26 | 492 | Along creek | 250 | 37° 22′ 29″ 28° 05′ 27″ |
| Total | - | 223 | 4779 | - | - | - |
All seeds were collected and kept separately in plastic bags for each mother tree and the bags were labelled. The number of families and the number of seedlings obtained for each population ranged from 16 to 27 and from 349 to 601, respectively.
Sowing of seeds was carried out in the Gokova Nursery of Mugla Province, Turkey in the spring of 2008. ENSO pots (Finland type) of 259 cm3 with 45 seedling capacities were used. Seedlings were grown under common conditions and were transplanted in the common garden test site in March 2009.
The experiment test site was located within the optimal distribution area of oriental sweet gum in Turkey (37° 00′ 18″ N latitude, 28° 30′ 14″ E longitude, elevation 100 m a.s.l. - Fig. 1). The test was established using a randomized complete block design in 25 blocks with single tree plot. Each block included 223 open pollinated (OP) families planted at a spacing of 3 × 4 m. The breast height diameter (mm), the height (cm) and the crown diameter (cm) of seedlings were measured at the age of five. The crown diameter of each tree was measured on two perpendicular directions and the two measurements were then averaged.
Statistical analyses
The initial height of seedlings at the time of plantation was used as covariate in the statistical model. Individual tree data were subjected to analysis of variance (ANOVA), following the statistical model below for all the traits studied (eqn. 1):
where Yijkl is the observation on the lth seedling, kth family, jth population, ith block, μ is the overall mean, A the regression coefficient, Xijkl is the establishment height of the seedlings (ijkl = 1, …, ≈5000), bi is the random effect of the ith block (i = 1, …, 25), pj the random effect of the jth populations (j=1, …, 9), fk(j) (k = 1,…, 27) the random effect of the kth family effect within the jth population, and eijkl is the experimental error.
Variance components were estimated by using REML (Restricted Maximum Likelihood) option of the PROC VARCOMP of the software SAS ver. 9.0 ([32]). Individual heritability, additive genetic coefficient of variation and genetic correlation were estimated by the following equations ([6], [9], [14] - eqn. 2, eqn. 3, eqn. 4):
where hi2 is the individual heritability, σf2 is the family variance, σe2 is the error variance, Cvg is the additive genetic coefficient of variation, XÌÂ is the general mean, rAxy is the genetic correlation, COV(x,y) is the covariance for x and y traits, σx2 is the variance for trait x and σy2 the variance for trait y. It was assumed that the open-pollinated seeds were half-sibs for the additive variance ([33]). Standard errors of individual heritability (eqn. 5) were derived from Dickerson’s method suggested by Dieters et al. ([10]) and the standard error of genetic correlation (eqn. 6) was derived by Falconer & Mackay ([14]), as follows:
where SE is the standard error, hx2 is the heritability for trait x and hy2 is the heritability for trait y.
Results
The highest mean of breast height diameter, height and crown diameter was found for KOYC, YILA, and YILA populations, respectively, though the lowest mean of all traits was observed for BOZD (Tab. 2). On the other hand, mean values of seedling from YATA population were also low of all traits, though higher than that observed for BOZD.
Tab. 2 - Means (± standard errors) of all traits for each population. (BHD): diameter at breast height; (H): height; (CD): crown diameter.
| Populations | BHD | H | CD |
|---|---|---|---|
| KOYC | 23.6 ± 0.3 | 245.9 ± 1.9 | 163.6 ± 1.6 |
| CETI | 23.2 ± 0.3 | 246.1 ± 2.0 | 165.3 ± 1.5 |
| YILA | 23.0 ± 0.3 | 250.6 ± 2.0 | 171.5 ± 1.6 |
| DEGI | 22.7 ± 0.3 | 248.6 ± 2.0 | 163.7 ± 1.5 |
| GUNL | 22.1 ± 0.3 | 243.8 ± 1.9 | 161.5 ± 1.5 |
| KIYR | 21.4 ± 0.3 | 235.6 ± 2.0 | 160.2 ± 1.6 |
| GUNN | 20.4 ± 0.3 | 237.3 ± 2.1 | 159.8 ± 1.7 |
| YATA | 18.0 ± 0.3 | 215.8 ± 2.1 | 150.7 ± 1.7 |
| BOZD | 15.7 ± 0.3 | 198.4 ± 2.2 | 128.4 ± 2.1 |
| Grand mean | 21. 5 ± 0.1 | 237.9 ± 0.7 | 159.8 ± 0.6 |
The variance components, the individual heritabilities and the additive genetic coefficient of variations for all traits of pooled data across populations are summarized in Tab. 3. The among-population variances were about 3 times higher than the family (within-population) variances for all the traits considered in this study. The individual heritability ranged from 0.11 (crown diameter) to 0.27 (height). The additive genetic coefficient of variation was the lowest for crown diameter and the highest for breast height diameter. All factors (block, population, family) were statistically significant.
Tab. 3 - Variance components, expressed as percentage of the total variation, individual heritability and additive genetic coefficient of variation for pooled data across populations. (BHD): diameter at breast height; (H): height; (CD): crown diameter; (***): P<0.0001; (a): IH (Initial height) used as covariate was statistically significant for all traits (P<0.0001).
| Sources of variation (a) |
Degrees of freedom |
Traits | ||
|---|---|---|---|---|
| BHD | H | CD | ||
| Block | 24 | 18.60*** | 20.09*** | 18.31*** |
| Pop | 8 | 11.82*** | 11.92*** | 9.69*** |
| Family(Pop) | 207 | 3.56*** | 4.60*** | 2.06*** |
| Error | 4336 | 66.02 | 63.39 | 69.94 |
| Total | 4767 | 100.00 | 100.00 | 100.00 |
| h i 2 | - | 0.21 ± 0.03 | 0.27 ± 0.04 | 0.11 ± 0.03 |
| Cvg (%) | - | 13.40 | 9.10 | 7.10 |
The analysis of variance of pooled data across populations revealed statistically significant differences both among populations and among families within populations.
The nine populations analyzed were divided into two groups, marginal and optimal, according to their geographic origin. Ozdilek et al. ([27]) also found that the optimum distribution area of oriental sweet gum could be considered as the genetic diversity center and the major refugium for the oriental sweet gum. The marginal group included the seedlings from YATA and BOZD populations, and the optimal group included the other 7 provenances.
Unlike optimal populations, marginal populations showed higher among-population variances compared to family variances. Mean values of breast height diameter, height and crown diameter in optimal group was found higher than those of marginal group. Individual heritability for breast height diameter, height and crown diameter of seedlings were 0.27 ± 0.10, 0.19 ± 0.08 and 019 ± 0.08 in marginal populations and 0.19 ± 0.04, 0.29 ± 0.05 and 0.09 ± 0.03 in the optimal group (Tab. 4). The additive genetic coefficient of variation for diameter at the breast height was higher than that of other traits for both marginal and optimal populations.
Tab. 4 - Variance components, expressed as percentage of the total variation, mean, individual heritability and additive genetic coefficient of variation for optimal and marginal populations. (BHD): diameter at breast height; (H): height; (CD): crown diameter; (***): P<0.0001; (a): IH (Initial height) is used as covariate and statistically significant for all traits (P<0.0001); (b): first degree of freedom is for optimal and second is for marginal populations; (c): ranges of traits are in the parenthesis.
| Sources of variation (a) |
Degrees of freedom (b) |
Optimal Populations | Marginal Populations | ||||
|---|---|---|---|---|---|---|---|
| BHD | H | CD | BHD | H | CD | ||
| Block | 24 | 21.4*** | 23.7*** | 20.9*** | 16.2*** | 16.5*** | 14.1*** |
| Pop | 6-1 | 2.0*** | 1.0*** | 0.9*** | 6.3*** | 7.1*** | 14.6*** |
| Family(Pop) | 170-37 | 3.7*** | 5.4*** | 1.8*** | 5.3*** | 3.6*** | 3.4*** |
| Error | 3559-720 | 72.9 | 69.9 | 76.4 | 72.2 | 72.8 | 67.9 |
| Total | 3903-806 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| Mean (c) | - | 23.3 (10-57) | 244.1 (72-421) | 163.7 (40-315) | 17.8 (8-48) | 248.6 (79-381) | 141.5 (30-260) |
| h i 2 | - | 0.19 ± 0.04 | 0.29 ± 0.05 | 0.09 ± 0.03 | 0.27 ± 0.10 | 0.19 ± 0.08 | 0.19 ± 0.08 |
| Cvg (%) | - | 12.80 | 9.10 | 6.20 | 16.70 | 8.30 | 10.80 |
Genetic and phenotypic correlations were slightly higher for the marginal populations than for the optimal ones (Tab. 5). The genetic correlations between breast height diameter and height were high and positive, but for the other trait pairs were low though positive.
Tab. 5 - Genetic (below diagonal) and phenotypic (above diagonal) correlations. (BHD): diameter at breast height; (H): height; (CD): crown diameter.
| Traits | Optimal Populations | Marginal Populations | ||||
|---|---|---|---|---|---|---|
| BHD | H | CD | BHD | H | CD | |
| BHD | - | 0.74 | 0.48 | - | 0.70 | 0.51 |
| H | 0.83 ± 0.04 | - | 0.45 | 1.00 ± 0.00 | - | 0.52 |
| CD | 0.45 ± 0.15 | 0.55 ± 0.08 | - | 0.52 ± 0.20 | 0.60 ± 0.19 | - |
Discussion
In this study, traits likely to be of adaptive importance, such as breast height diameter, height and crown diameter, were analyzed in 5-year-old seedlings of Liquidambar orientalis grown in a common garden experiment. Such traits may be relevant in the competition for light and other resources in forest trees. Vitasse et al. ([37]) emphasized that under favorable conditions, a higher growth rate increases species’ competitive ability, survival, and long-term success. Thus, it was assumed that breast height diameter, height, and crown diameter, measured on about 5000 seedlings in the common garden test, could be used to assess the level of adaptation of oriental sweet gum.
For all the traits considered in this study, mean values of seedlings from marginal populations (BOZD and YATA) were lower than the general means, while those observed for optimal provenances (KOYC, GUNL, DEGI, YILA, KIYR, and CETI) were higher, except for GUNN (Tab. 2). Moreover, mean values of breast height diameter, height and crown diameter in the optimal group were higher by 4.5 mm, 35.5 cm and 22.2 cm, respectively, than those observed in the marginal group (Tab. 4). Vitasse et al. ([37]) also found that populations from low altitudes (optimal) had higher growth rates than populations from high altitudes (marginal). Considering the distribution range of oriental sweet gum ([2]), optimal group populations are located in sites of favorable growing conditions and in the genetic diversity center of the species ([27]). Contrastingly, the marginal group of populations lives in sites of peripheral or marginal growing conditions which expose trees to a stressful environment. Therefore, the observed differences in growth rate between optimal and marginal populations might be sourced from their growth conditions.
High variation in quantitative traits of oriental sweet gum was found among populations and among families within populations. The population variances were about 3-4 times higher than the family variances for all the traits of the pooled data across populations. The same trend existed also in marginal populations. Velioglu et al. ([36]) found a low gene flow coefficient between populations (number of migrants Nm = 0.42) and a high genetic divergence among populations (genetic distance GST = 0.54). Not only the pooled data across all populations but also findings of the marginal populations supported Velioglu et al. ([36]) on the population differentiation. Unlike marginal populations, the family variances in optimal provenances were about 2-5 higher than among-populations variances. Baliuckas et al. ([4]) suggest that a large additive variance within populations for adaptive traits is the best assurance for future adaptation. On the other hand, Jensen & Hansen ([18]) suggested that the larger among-population variation (compared to that of families) observed in Quercus robur and Q. petrea reflects a strong adaptation to local sites. Therefore, in the light of the above considerations, both optimal and marginal population groups are functional in maintaining the adaptability of oriental sweetgum to future changes.
Individual heritabilities (0.11-0.27) of pooled data across all populations were in accordance with Cornelius ([9]) who reviewed heritabilities of 67 forest tree species and recorded a range of 0.1-0.3. A similar trend was observed for the heritabilities of optimal and marginal populations (0.09-0.29). Moreover, heritabilities for optimal populations were more similar to those calculated from pooled data than those of marginal populations. Thus, marginal and optimal populations were different in terms of heritability. While breast height diameter and crown diameter were more heritable for marginal populations, height was more heritable for optimal populations. On the other hand, all heritability values estimated in this study could be biased as obtained from a single trial experiment ([38]). In this context, considering a possible overestimation of heritability, oriental sweet gum exhibited heritability values similar to those observed for other forest trees when all populations were studied together, though those values were different when marginal and optimal populations were analyzed separately. Especially, heritability of crown diameter in marginal populations was two times higher than that of optimal populations.
The additive genetic coefficient of variation in breast height was the highest, when compared to that estimated for the other traits, in both the marginal and optimal populations. Cornelius ([9]) reported that the additive genetic coefficient of variation in forest trees was less than 15% on average. The additive genetic coefficient of variation is a good indicator of long-term evolution of a trait and suggests that the studied populations have a good potential for adaptation under changing environmental conditions ([16], [12], [39], [5]). In this context, breast height diameter in oriental sweet gum might be considered a more plastic trait. On the other hand, the genetic coefficient of variation for the crown diameter in marginal populations was higher than that estimated for optimal provenances. While the mean crown diameter of seedlings from optimal populations ranged from 159.8 cm to 171.5 cm, mean crown diameter in YATA (the northernmost population) was 150.7 cm and 128.4 cm in BOZD (at the highest elevation population - Tab. 2). Because of the narrowest crown diameter recorded in BOZD population, it could be hypothesized that the crown diameter might be related to adaptation of the species to harsh environmental conditions like snow.
The genetic correlation in marginal populations was slightly higher than the genetic correlations in optimal populations. Genetic correlations of breast height diameter and height were high and positive as reported for most forest trees. This suggests that same sets of genes regulate the height and the breast height diameter traits for both marginal and optimal populations ([12]).
Oriental sweet gum is one of the 110 species determined by EUFORGEN to be the subject of studies related to genetic conservation ([2], [13]). Our findings on oriental sweet gum populations can be evaluated from the perspective of genetic conservation of forest genetic resources under the current climate change. The latest projections indicate that summer temperatures (June-August) will increase by 3-4 °C in most parts of Europe by 2081 to 2100, and even 4-5 °C in some Mediterranean regions ([17]) where the oriental sweet gum is distributed. The effects of climate change on forest genetic resources will mostly depend on the size and the distribution of the current tree populations and on the biology of the species. A widespread species is expected to be less threatened by the impacts of climate change in comparison to a species with a narrow distribution and small populations ([20]). On the other hand, Kreyling et al. ([21]) emphasized on local adaptations to environmental conditions and suggested that the increased environmental stress (warming, extreme drought) due to climate change, in combination with the decrease of genetic mixing due to isolation, may lead to stronger local adaptations in geographically marginal populations. Kelleher et al. ([20]) suggested two predictions on climate change and forest gene resources which refer also to the oriental sweet gum. The first prediction was that “southern species will face increasing population fragmentation and reduction in population numbers. Some of the southernmost populations are likely to become extinct, resulting in an overall reduction in forest genetic resources.” The second was: “within the core of the distribution the most likely outcome will be a change in the species composition through changes in forest dynamics with a possible reduction in genetic diversity”. Conversion to farm and settlement pressure (tourism, road building, etc.) were the reasons for fragmentation of oriental sweet gum. As a result of these threats, its distribution area decreased by about a half ([2]). In the existing conditions, optimal populations are expected to be subject to fragmentation resulting from land conversion to farms, building roads etc. Regarding the first prediction by Kelleher et al. ([20]), optimal populations should be protected against fragmentation. In this context, at least some of the optimal populations which are far away from human settlements can be designated as in situ genetic conservation units. On the other hand, researches of assisted migration on oriental sweet gum can be carried out to mitigate the second prediction of Kelleher et al. ([20]). Marginal populations have shown differentiation, but their genetic plasticity (in terms of additive genetic coefficient of variation) was similar to the optimal ones. Richter et al. ([30]) stated that autochthonous provenances of Pinus sylvestris L. had the potential for resistance to changes in climatic conditions as a function of both phenotypic plasticity and genotypic variation. Regarding the European perspective on genetic conservation, the marginal populations of oriental sweet gum may be also thought as an integral part of a strategy for the conservation of its adaptability and evolutionary potential to face the current climate change.
This study has analyzed nine populations of oriental sweet gum, of which two from marginal and seven from optimal sites for the species. As a consequence, standard errors of heritability and genetic correlation in the marginal group were higher than in the optimal group of populations. For this reason, the estimation of genetic parameters for marginal populations should be confirmed using larger datasets. Moreover, heritability was assessed on young seedlings (5-year-old) and therefore predictions of expressed genetic variation in adult trees must be taken with caution. Finally, several common garden experiments established at different localities are needed to assess genotype-environment interactions and confirm the conclusions of this study. Nonetheless, as no previous studies on quantitative traits for oriental sweet gum do exists, the results from marginal and optimal groups are thought to supply an adequate information for the genetic conservation of the species.
Conclusions
Known patterns of genetic variation among populations are useful in gene resource management as a guide for seed transfer and genetic conservation. In fragmented habitats, opportunities for migration are limited and the conservation of evolutionary potential as a basis for facing environmental change is likely to become especially important. Significant differences were found for each trait among populations of oriental sweet gum. In pooled data across populations and marginal populations, among-population variation was higher than within-population variation for all the traits. Regarding marginal populations, the large between-population variation may also reflect a strong adaptation to local sites, which justify a high conservation priority and the establishment of in situ gene conservation units. On the other hand, the family variances were substantially higher than populations variances in provenances from optimal sites, which provide a good prospect for coping with changed environmental conditions. The ongoing deforestation rate in the optimal distribution area of the species, mainly due to the conversion of natural stands to farmlands, calls for urgent conservation actions for the remnant forest genetic resources.
The presence of additive genetic variation for adaptive traits is important for populations to adapt to a changing environment. The pattern of heritabilities and the genetic coefficients of variations were different, but the pattern of genetic correlations was alike between marginal and optimal populations. The breast height diameter might be the most plastic trait in oriental sweet gum as revealed by highest genetic coefficients of variation in both marginal and optimal groups.
The inclusion of oriental sweet gum in EUFORGEN allow to apply the European perspective on gene conservation to the species. Furthermore, assisted migration can be considered for oriental sweet gum to cope with current climate change, but extensive research is needed on this topic.
Acknowledgments
The research Project on Oriental sweet gum has been carried out by the Forest Tree Seeds and Tree Breeding Research Institute Directorate in Ankara/Turkey since 2009. The number of the Project is ANK033 1624/2009-2026.
Many thanks to Ercan Velioglu, Turgay Ezen, Sadi Siklar and Hikmet Öztürk for their valuable contributions.
References
Gscholar
CrossRef | Gscholar
Gscholar
Gscholar
Gscholar
Online | Gscholar
CrossRef | Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Karabuk University, Faculty of Forestry, Karabuk (Turkey)
Corresponding author
Paper Info
Citation
Alan M (2018). Comparison of genetic parameters between optimal and marginal populations of oriental sweet gum on adaptive traits. iForest 11: 510-516. - doi: 10.3832/ifor2450-011
Academic Editor
Fulvio Ducci
Paper history
Received: Apr 04, 2017
Accepted: May 15, 2018
First online: Jul 18, 2018
Publication Date: Aug 31, 2018
Publication Time: 2.13 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2018
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 47798
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 39657
Abstract Page Views: 3105
PDF Downloads: 3972
Citation/Reference Downloads: 5
XML Downloads: 1059
Web Metrics
Days since publication: 2752
Overall contacts: 47798
Avg. contacts per week: 121.58
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
(No citations were found up to date. Please come back later)
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Age trends in genetic parameters for growth and quality traits in Abies alba
vol. 9, pp. 954-959 (online: 07 July 2016)
Research Articles
Genetic variation of Fraxinus excelsior half-sib families in response to ash dieback disease following simulated spring frost and summer drought treatments
vol. 9, pp. 12-22 (online: 08 September 2015)
Technical Reports
Population genetic structure of Platanus orientalis L. in Bulgaria
vol. 4, pp. 186-189 (online: 11 August 2011)
Commentaries & Perspectives
The genetic consequences of habitat fragmentation: the case of forests
vol. 2, pp. 75-76 (online: 10 June 2009)
Research Articles
Comparison of range-wide chloroplast microsatellite and needle trait variation patterns in Pinus mugo Turra (dwarf mountain pine)
vol. 10, pp. 250-258 (online: 11 February 2017)
Technical Advances
Gene flow in poplar - experiments, analysis and modeling to prevent transgene outcrossing
vol. 5, pp. 147-152 (online: 13 June 2012)
Review Papers
Genetic diversity and forest reproductive material - from seed source selection to planting
vol. 9, pp. 801-812 (online: 13 June 2016)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword