Preliminary study on genetic variation of growth traits and wood properties and superior clones selection of Populus ussuriensis Kom.
iForest - Biogeosciences and Forestry, Volume 12, Issue 5, Pages 459-466 (2019)
doi: https://doi.org/10.3832/ifor2991-012
Published: Sep 29, 2019 - Copyright © 2019 SISEF
Research Articles
Abstract
In this study the genetic variation of growth traits and wood properties was assessed in 45 clones of Populus ussuriensis Kom. grown in a 10-year-old experimental forest located in Northeastern China and a preliminary selection of superior clones was performed based on multi-trait selection index. The following traits were analyzed: tree height, H; diameter at breast height, DBH; volume, V; basic wood density, BWD; fiber length, FL; fiber length-width ratio, FL/W; microfibril angle, MA; cell wall percentage, CWP; fiber double wall thickness, FWT; vessel proportion, VP; wood fiber proportion, FP; wood ray proportion, RP; holocellulose content, HC. The results revealed significant differences (P<0.05) in DBH, V, BWD, FWT, VP and FP among different clones. Broad-sense heritabilities for growth traits and wood properties ranged from 0.020 to 0.749, therefore growth traits and wood properties are moderately or weakly controlled. Negative correlations between growth traits and most wood properties were also found. According to the multi-trait selection index at a selected rate of 10% and survival rates, 3 superior clones (I18, H16, C13) were selected, and the DBH, V, BWD, FL and FP of superior clones were higher than those of all clones by 2.83%, 9.81%, 3.40%, 6.59% and 0.31%, the MA, FWT, VP were lower than those of all clones by 7.54%, 0.39%, 2.12%, respectively. These superior clones could be used as starting breeding material for P. ussuriensis.
Keywords
Populus ussuriensis Kom., Growth Traits, Wood Properties, Genetic Variation, Multi-trait Index Selection
Introduction
Forests are the largest terrestrial ecosystem on the earth and are an important part of the global biosphere. Forests are also the gene pool and energy bank for earth, and play a vital role in maintaining the ecological balance of the whole earth. In particular, forest ecosystems constitute approximately 82% of the continental biomass and comprise more than 50% of terrestrial biodiversity ([30], [28]). According to FAO ([13]), the estimate of forest cover on terrestrial surface in 2015 was 30.6%, and between 1990 and 2015 the deforestation rate has slowed down and the afforestation has increased globally. Plantations have provided more goods and services than those derived from natural forests. Indeed, the area covered by mature forests decreased since 2000 ([33]). Whether they can continue to provide enough goods and services is a worry. In contrast, the demand for industrial wood and firewood is increasing. With the implementation of natural forest protection projects in China, wood products such as timber are now mainly dependent on plantations. Hence, there is an urgent need for fast-growing, high-quality plantations.
Populus ussuriensis Kom. has a strong adaptability and very high yield. It is also suitable for adjusting stand structure in commercial forest plantations ([36]). In addition, it is one of the main tree species of forest renewal in the eastern mountainous areas of Northeastern China. P. ussuriensis is widely distributed in Northern China, the Korean Peninsula and the Far East ([45]). It grows mainly in the mountains at an altitude of 300-1200 m a.s.l. on slightly acidic brown forest soil or mountain dark brown soil. P. ussuriensis has straight trunk and the durability of white timber, and it is an important resource for construction, paper and plywood. Zhang et al. ([42]) studied the fiber morphology and basic density of P. ussuriensis, finding that it can meet the requirements of pulpwood. Furthermore, Wang et al. ([37]) found significant differences in H, DBH and V among clones of P. ussuriensis.
The properties of poplar wood have a deep effect on its industrial use. Indeed, large differences in wood properties has been found among different poplar clones, between different individuals of the same clone and also between different parts of the same individual tree. Such differences are largely influenced by site conditions, cultivation measures and other factors in a complex way. Up to now, studies about the physical properties of poplar wood mainly focused on wood density, moisture content and shrinkage rate. For example, Li & Xu ([22]) found a significant linear positive correlation between wood density at breast height and the density of the whole tree in the poplar clone I-63. Xing & Zhang ([38]) studied triploid Chinese white poplar and showed that arid shrinkage rate of wood was significantly different between clones and strongly controlled by genes, with variation patterns in the longitudinal direction decreasing with the increment of tree height. Yanchuk et al. ([40]) showed a slight negative phenotypic correlation between wood density and growth rate in trembling aspen (Populus tremuloides Michx.), as well as a slight positive phenotypic correlation between fiber length and growth rate. In addition, large differences in mechanical and chemical properties of wood among different poplar varieties and clones have been reported by Wang et al. ([35]), resulting in the selection of the best varieties and clones for use in the short-period timber industry.
The above studies demonstrated the existence of significant differences in anatomical, physical, mechanical and chemical properties of poplar wood among species and clones. However, a few studies are currently available in the literature on wood variability of P. ussuriensis, in particular on wood properties such as microfibril angle, cell wall percentage, fiber double wall thickness and tissue proportion.
Selection is a powerful tool for genetic improvement of forest trees and the selection method has a relevant effect on genotype improvement. One of the major difficulties faced by tree breeders is the negative correlation between wood properties and growth traits when selecting promising genotypes ([26]). The selection index is one of the helpful tools in the selection process, as it allows for multiple traits of interest to be selected simultaneously ([34]). Missanjo & Matsumura ([26]) used a multiple trait selection index to select superior trees of Khasi pine (Pinus kesiya Royle ex Gordon).
In this study, clones were collected from the Chinese range of P. ussuriensis and planted at the Maoershan Experimental Forest Farm of Northeast Forestry University (Heilongjiang, China - Fig. 1). Ten years after planting, the growth traits and wood properties of each clone were determined. The goals were to evaluate variation patterns in growth traits and wood properties of different P. ussuriensis clones and preliminary selection of fast-growing, high-quality clones for pulpwood.
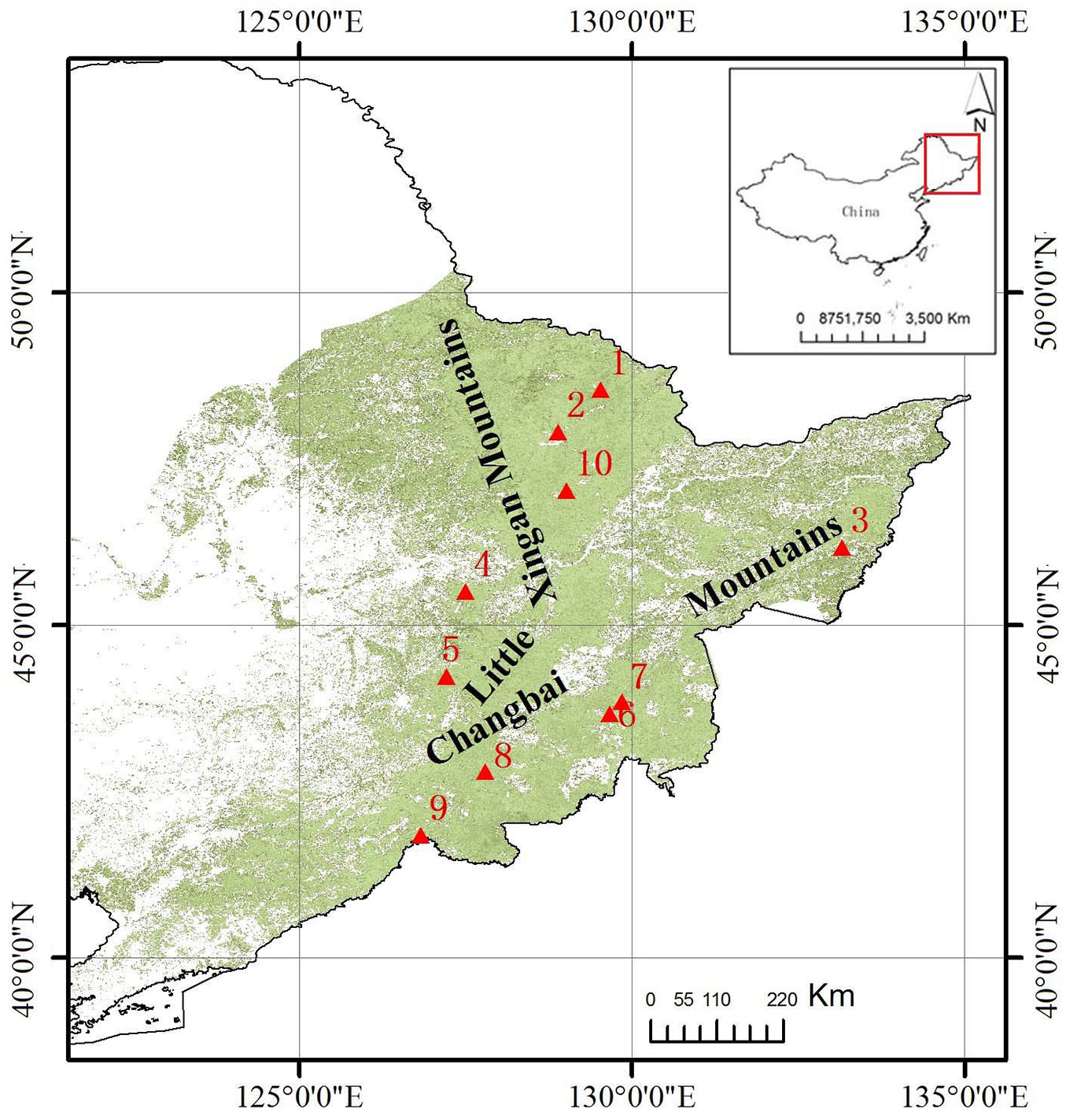
Fig. 1 - The natural range of P. ussuriensis in Northeastern China (in green) and the original sampling locations of clones (red triangles). For each location, the site name and the clones collected therein are listed below. (1): Xinqing - D9, F6; (2): Yichun - E5, E8, E9; (3): Dongfanghong - J9, J19, J20; (4): Maoershan - M10, M13, M14, M15, M16, M17, M19, M20; (5): Shangying - D16, D17; (6): Jiangshanjiao - H4, H5, H7, H8, H12, H13, H16; (7): Wangqing - D13, I5, I8, I10, I18; (8): Lushuihe - B5, B6, B8, D5, D11; (9): Linjiang - C1, C9, C12, C13, C14, C15, C16; (10): Dailing - D22, D23, D24. Maoershan (4) is the experimental site at the Northeast Forestry University (Heilongjiang).
Materials and methods
Materials
The materials used in this study were 45 P. ussuriensis clones (Tab. 1). In the winter of 2001-2002, cuttings were collected from excellent trees within the Chinese distribution range of P. ussuriensis. In 2003, cuttings were propagated in the nursery and then planted the field in 2007 at an experimental forest site in Maoershan, Shangzhi City, Heilongjiang Province (North-East China). The trial was conducted under a randomized complete block design consisting of three blocks, with 45 clones in every block; 15 trees of each clone were planted in one block, spaced at 2 × 2 m. Unfortunately, some trees died due to pests and diseases.
Tab. 1 - The survival rates of 45 poplar (P. ussuriensis) clones analyzed in this study.
| Clone | Survival rate (%) |
Clone | Survival rate (%) |
Clone | Survival rate (%) |
|---|---|---|---|---|---|
| I10 | 95.24 | F6 | 80.95 | J20 | 73.81 |
| B5 | 92.86 | H13 | 80.95 | C9 | 69.05 |
| B8 | 92.86 | H5 | 80.95 | D11 | 69.05 |
| M20 | 92.86 | H7 | 80.95 | I5 | 69.05 |
| M19 | 90.48 | J9 | 80.95 | D23 | 64.29 |
| C14 | 88.10 | C13 | 78.57 | E9 | 64.29 |
| M14 | 88.10 | D13 | 78.57 | H12 | 64.29 |
| C16 | 85.71 | D22 | 78.57 | C15 | 61.90 |
| D24 | 85.71 | M15 | 78.57 | I8 | 59.52 |
| M10 | 85.71 | I18 | 76.79 | C12 | 57.14 |
| M17 | 85.71 | B6 | 76.19 | H4 | 57.14 |
| D16 | 83.33 | M13 | 76.19 | E5 | 52.38 |
| D17 | 83.33 | M16 | 76.19 | J19 | 52.38 |
| H8 | 83.33 | D5 | 73.81 | C1 | 47.62 |
| D9 | 80.95 | H16 | 73.81 | E8 | 47.62 |
Determination of growth traits
During the autumn of 2015 (10 years after planting), tree height (H) and diameter at breast height (DBH) of all trees were measured. The stem volume (V) of each tree was calculated based on DBH (D1.3) and H, as described by Li & Li ([23] - eqn. 1):
Determination of wood properties
Wood cores were taken at DBH along the north-south direction from three ramets per clone in every block, and used for the assessment of subsequent wood properties.
The basic wood density (BWD) was measured using the saturated water method ([41]).
The fiber lengths and widths were measured by the Jeffrey method ([31]), with all materials soaking in a mixture of 10% nitric acid and 10% chromic acid for 4 hours. Materials were rinsed with deionized water several times until the solution was neutral. Samples were measured under a Zeiss™ optical microscope, and 50 fibers were measured for each sample.
Microfibril angles (MA) were determined by the iodine staining method ([9]). The wood cores were sliced in the middle of the annual rings near the bark, with a thickness of approximately 15-19 μm per slice. Angles were observed under a 400-fold readable angle microscope, and 20 microfibril angles were measured for each sample.
Measurements of cell wall percentage (CWP), fiber double wall thickness (FWT) and tissue proportion were also taken. Softened wood samples were sliced using a slippery wood slicer, with slices taken at a thickness of 12-16 μm. After dyeing, dehydration by ethanol and clearing by xylene, a Nikon™ 80i® microscope and a DS-Ri1® digital camera (Nikon Imaging Inc., Tokyo, Japan) were used to collect clear wood photos in the middle of the two rounds near the bark according to measurement requirements ([44]). TDY-5.2 color image computer analysis software was used for image analysis.
Holocellulose content (HC) was measured using an ANKOM 2000i® automated fiber analyzer (ANKOM Technology, Macedon, NY, USA).
Statistical analysis
Analysis of variance (ANOVA) was performed using the statistical package SPSS® ver. 22.0 (IBM, Armonk, NY, USA). The following linear models were used (eqn. 2):
where Yij is the observations, μ is the mean, Pi is the clone effect, Bj is the block effect, and εij is the random error.
The genotypic (σ2g) and phenotypic (σ2p) variances were calculated using the mean squares from ANOVA as described by Metougui et al. ([25] - eqn. 3, eqn. 4):
where A = σ2ε +rσ2g is the mean square between clones, B = σ2ε, is the mean square within families, and r is the number of replications.
The phenotypic coefficient of variation (PCV, %) and the genotypic coefficient of variation (GCV, %) were estimated as follows ([18] - eqn. 5, eqn. 6):
where {bar} X is the mean value of a single trait among all clones. Broad-sense heritability (H2) for the clones was calculated using the following equation ([39] - eqn. 7):
where F is the F-value from the ANOVA table.
The genetic correlation coefficient was calculated as ([3] - eqn. 8):
where σ2g(xy) is the genetic covariance of trait x and trait y, σ2g(x) is the genetic variance of trait x, and σ2g(y) is the genetic variance of trait y.
The selection index was obtained using the software package DPS ver. 16.05 (DPS Software Ltd., Enfield, UK). The index value (I) is the sum of the weighted values for each trait under consideration ([21] - eqn. 9):
where bi is the index coefficient for trait i and xi is the average of trait i.
The selection index coefficient was calculated as ([26] - eqn. 10):
where b is the weighting factor column vector, P is the phenotypic variance matrix, A is the genetic variance matrix, and W is the economic weight column vector.
Results
Survival rates of clones
The average survival rate of the 45 P. ussuriensis clones analysed in this study was 75.46%. The survival rates of clone I10, B5, B8, M20 and M19 were higher than 90%. Some clones have low survival rates, e.g., clone C1 and E8 were lower than 50%.
Variation and genetic parameters
The variation and genetic parameters of growth traits and wood properties are shown in Tab. 2. Total average H was 10.34 m, with the highest clone B6 (12.44 m) higher than the lowest clone E5 (7.77 m) by 58%. Total average DBH of all clones was 9.32 cm, with the highest clone D23 (10.76 cm) higher than the lowest clone E5 (6.58 m) by 64%. Total average V was 0.0375 m3, with the highest clone B6 (0.0537 m3) higher than the lowest clone E5 (0.0175 m3) by 207%. PCVs and GCVs of growth traits of all clones varied from 12.83% to 31.55% and 2.70% to 14.61%, respectively. H2 of growth traits varied from 0.084 to 0.663.
Tab. 2 - Variation and genetic parameters of growth traits and wood properties. (H): tree height, m; (DBH): diameter at breast height, cm; (V): volume, m3; (BWD): basic wood density, g/cm3; (FL): fiber length, µm; (FL/W): fiber length-width ratio; (MA): microfibril angle, °; (CWP): cell wall percentage, %; (FWT): fiber double wall thickness, µm; (VP): vessel proportion, %; (FP): wood fiber proportion, %; (RP): wood ray proportion, %; (HC): holocellulose content, %; (PCV): phenotypic coefficient of variation, %; (GCV): genotypic coefficient of variation, %.
| Traits | Average | Range | PCV | GCV | H 2 | σ 2 g | σ 2 p |
|---|---|---|---|---|---|---|---|
| H | 10.34 | 7.77-12.44 | 15.68 | 2.70 | 0.084 | 0.08 | 2.63 |
| DBH | 9.32 | 6.58-10.76 | 12.83 | 8.07 | 0.663 | 0.57 | 1.43 |
| V | 0.0375 | 0.0175-0.0537 | 31.55 | 14.61 | 0.477 | 0.00003 | 0.0001 |
| BWD | 0.2705 | 0.2421-0.3083 | 7.39 | 5.23 | 0.749 | 0.0002 | 0.0004 |
| FL | 767.65 | 677.01-855.56 | 9.21 | 2.75 | 0.227 | 445.78 | 4998.28 |
| FL/W | 29.52 | 26.96-35.04 | 8.33 | 2.71 | 0.263 | 0.64 | 6.68 |
| MA | 12.63 | 9.30-18.67 | 20.49 | 7.30 | 0.302 | 0.85 | 6.70 |
| CWP | 38.00 | 32.13-46.87 | 13.00 | 4.83 | 0.325 | 3.37 | 24.39 |
| FWT | 3.96 | 3.39-5.06 | 12.88 | 7.14 | 0.557 | 0.08 | 0.26 |
| VP | 25.99 | 20.35-32.38 | 14.28 | 8.31 | 0.606 | 4.66 | 13.77 |
| FP | 69.57 | 63.57-75.45 | 5.21 | 3.06 | 0.614 | 4.54 | 13.12 |
| RP | 4.45 | 3.45-5.45 | 17.26 | 5.02 | 0.220 | 0.05 | 0.59 |
| HC | 75.46 | 70.05-80.11 | 3.29 | 1.07 | 0.261 | 0.65 | 6.18 |
Total average BWD was 0.2705 g cm-3, with the biggest clone M17 (0.3083 g cm-3) bigger than the smallest clone D16 (0.2421 g cm-3) by 27%. Total average FL was 767.65 µm, with the longest clone C14 (677.01 µm) longer than the smallest clone I18 (855.56 µm) by 26%. Total average FL/W, CWP and FWT were 29.52, 38.00% and 3.96 µm, respectively. The highest average FL/W (35.04), CWP (46.87%) and FWT (5.06 µm) were observed for clone J19 and were higher than the lowest average FL/W, CWP and FWT in clone C14 (26.96), M14 (32.13%) and D13 (3.39 µm) by 30%, 46% and 49%, respectively. Total average MA was 12.63°, with the highest clone J9 (18.07°) higher than the lowest clone I8 (9.30°) by 94 %. Total average VP was 25.99%, with the highest clone D17 (32.38%) higher than the lowest clone C1 (20.35%) by 59 %. Total average FP was 69.57%, with the highest clone E9 (75.45%) higher than the lowest clone D17 (63.57%) by 19%. Total average RP was 4.45%, with the highest clone B8 (5.45%) higher than the lowest clone E9 (3.45%) by 58%. Total average HC was 75.46%, with the highest clone E8 (80.11%) higher than the lowest clone D13 (70.05%) by 14 %. PCVs and GCVs of wood properties for all clones varied from 3.29% to 20.49% and 1.07% to 8.31%, respectively. H2 of wood properties varied from 0.220 to 0.749. BWD, FWT, VP and FP showed moderate genetic control, while all of the wood properties were genetically controlled to at least a mild degree.
ANOVA for growth traits and wood properties
The ANOVA results for growth traits and wood properties for 10-year-old clones of P. ussuriensis are shown in Tab. 3. The results showed significant (P<0.05) differences in DBH, V, BWD, FWT, VP and FP among clones. In addition, there were significant (P<0.05) differences for all growth traits and wood properties among blocks, except for the MA and HC traits.
Tab. 3 - Results of the ANOVA on growth traits and wood properties. (H): tree height; (DBH): diameter at breast height; (V): volume; (BWD): basic wood density; (FL): fiber length; (FL/W): fiber length-width ratio; (MA): microfibril angle; (CWP): cell wall percentage; (FWT): fiber double wall thickness; (VP): vessel proportion; (FP): wood fiber proportion; (RP): wood ray proportion; (HC): holocellulose content; (SV): sources of variation; (*): p< 0.05; (**): p< 0.01.
| Variable | SV | df | MS | F |
|---|---|---|---|---|
| H | Clone | 44 | 2.787 | 1.092 |
| Block | 2 | 36.398 | 14.260** | |
| Error | 88 | 2.552 | - | |
| DBH | Clone | 44 | 2.559 | 2.967** |
| Block | 2 | 30.688 | 35.577** | |
| Error | 88 | 0.863 | - | |
| V | Clone | 44 | 0.000 | 1.912** |
| Block | 2 | 0.003 | 27.276** | |
| Error | 88 | 0.000 | - | |
| BWD | Clone | 44 | 0.001 | 3.984** |
| Block | 2 | 0.003 | 12.307** | |
| Error | 88 | 0.000 | - | |
| FL | Clone | 44 | 5889.838 | 1.294 |
| Block | 2 | 52237.796 | 11.475** | |
| Error | 88 | 4552.502 | - | |
| FL/W | Clone | 44 | 7.319 | 1.357 |
| Block | 2 | 63.509 | 11.774** | |
| Error | 88 | 5.394 | - | |
| MA | Clone | 44 | 8.387 | 1.433 |
| Block | 2 | 5.084 | 0.869 | |
| Error | 88 | 5.851 | - | |
| CWP | Clone | 44 | 31.139 | 1.482 |
| Block | 2 | 293.697 | 13.976** | |
| Error | 88 | 21.015 | - | |
| FWT | Clone | 44 | 0.409 | 2.255** |
| Block | 2 | 7.053 | 38.886** | |
| Error | 88 | 0.181 | - | |
| VP | Clone | 44 | 23.103 | 2.535** |
| Block | 2 | 74.965 | 8.227** | |
| Error | 88 | 9.112 | - | |
| FP | Clone | 44 | 22.204 | 2.588** |
| Block | 2 | 27.390 | 3.193* | |
| Error | 88 | 8.579 | - | |
| RP | Clone | 44 | 0.689 | 1.282 |
| Block | 2 | 15.135 | 28.184** | |
| Error | 88 | 0.537 | - | |
| HC | Clone | 44 | 7.487 | 1.354 |
| Block | 2 | 0.315 | 0.057 | |
| Error | 88 | 5.531 | - |
Relationships among traits
The relationships among traits are shown in Tab. 4. Both H and DBH showed significant positive correlations (P<0.01) with V. The phenotypic and genetic correlation coefficients were 0.887 and 0.990 for H and 0.900 and 0.980 for DBH, respectively. H had a significant positive correlation (P <0.01) with DBH, with a phenotypic and genetic correlation coefficient of 0.690 and 0.990, respectively.
Tab. 4 - Correlation coefficients of different traits. The phenotypic correlation coefficient are shown above the diagonal (upper right part), while the genetic correlation coefficient are reported below the diagonal (lower left part). (H): tree height; (DBH): diameter at breast height; (V): volume; (BWD): basic wood density; (FL): fiber length; (FL/W): fiber length/width; (MA): microfibril angle; (CWP): cell wall percentage; (FWT): fiber double wall thickness; (VP): vessel proportion; (FP): wood fiber proportion; (RP): wood ray proportion; (HC): holocellulose content; (*): p< 0.05; (**): p< 0.01.
| - | H | DBH | V | BWD | FL | FL/W | MA | CWP | FWT | VP | FP | RP | HC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | 1 | 0.690** | 0.887** | -0.375** | 0.169* | -0.099 | -0.050 | -0.043 | 0.133 | 0.064 | -0.085 | 0.072 | 0.050 |
| DBH | 0.990 | 1 | 0.900** | -0.505** | 0.361** | -0.045 | 0.009 | -0.117 | 0.103 | -0.219* | 0.190* | 0.165 | 0.087 |
| V | 0.990 | 0.980 | 1 | -0.474** | 0.328** | -0.065 | 0.009 | -0.085 | 0.121 | -0.139 | 0.108 | 0.157 | 0.113 |
| BWD | -0.987* | -0.670 | -0.400 | 1 | -0.221** | 0.177* | 0.051 | 0.162 | 0.004 | -0.023 | 0.024 | -0.001 | -0.062 |
| FL | 0.980 | 0.828 | 0.980 | 0.135 | 1 | 0.621** | -0.064 | 0.066 | 0.302** | -0.211* | 0.152 | 0.281** | 0.061 |
| FL/W | -0.203 | -0.338 | -0.166 | 0.789 | 0.113 | 1 | -0.067 | 0.331** | 0.393** | -0.117 | 0.046 | 0.313** | -0.051 |
| MA | 0.035 | 0.068 | 0.100 | 0.265 | -0.047 | 0.393 | 1 | -0.025 | -0.086 | -0.129 | 0.127 | 0.034 | -0.141 |
| CWP | -0.999 | -0.572 | -0.857 | 0.503 | -0.280 | 0.998 | 0.461 | 1 | 0.787** | 0.030 | -0.120 | 0.362** | -0.097 |
| FWT | -0.995 | -0.435 | -0.658 | 0.352 | 0.314 | 0.970 | 0.174 | 0.970 | 1 | -0.106 | 0.010 | 0.412** | 0.033 |
| VP | 0.999 | -0.121 | -0.104 | -0.107 | -0.574 | -0.171 | -0.945* | -0.035 | 0.088 | 1 | -0.972** | -0.314** | -0.109 |
| FP | -0.940* | 0.164 | 0.153 | 0.054 | 0.597 | 0.233 | 0.892* | 0.047 | -0.085 | -0.995** | 1 | 0.084 | 0.115 |
| RP | -0.625 | -0.388 | -0.437 | 0.517 | -0.140 | -0.574 | 0.625 | -0.118 | -0.042 | -0.175 | -0.241 | 1 | -0.004 |
| HC | 0.468 | 0.091 | 0.379 | 0.414 | 0.990 | 0.865 | 0.150 | 0.990 | 0.640 | 0.398 | -0.241 | -0.241 | 1 |
BWD had a significant positive correlation (P<0.05) with FL/W and a significant negative correlation (P<0.01) with FL. The phenotypic correlation coefficients (the corresponding genetic correlation coefficients are reported in brackets henceforth) were 0.117 (0.789) and -0.221 (0.135) for FL/W and FL respectively. FL had a significant positive correlation (P<0.01) with FL/W, FWT and RP and a significant negative correlation (P<0.05) with VP. The phenotypic (genetic) correlation coefficients were 0.621 (0.113), 0.302 (0.314), 0.281 (-0.140) and -0.211 (-0.574) for these traits, respectively. FL/W had significant positive correlations (P<0.01) with CWP, FWT and RP. The phenotypic (genetic) correlation coefficients were 0.331 (0.998), 0.393 (0.970) and 0.313 (0.233) for these traits, respectively. CWP had significant positive correlations (P<0.01) with FWT and RP. The phenotypic (genetic) correlation coefficients were 0.787 (0.970) and 0.362 (-0.118) for FWT and RP, respectively. FWT showed a significant positive correlation (P<0.01) with RP. The phenotypic (genetic) correlation coefficient was 0.412 (-0.042). VP had significant negative correlations (P<0.01) with FP and RP. The phenotypic (genetic) correlation coefficients were -0.972 (-0.995) and -0.314 (-0.175) for these traits, respectively.
DBH had a significant negative correlation (P<0.05) with VP and a significant positive correlation (P<0.05) with FP. The phenotypic (genetic) correlation coefficients were -0.219 (-0.121) and 0.190 (0.164) for these traits, respectively. BWD had significant negative correlations (P<0.01) with H, DBH and V. There were negative correlations between growth traits and BWD. The phenotypic (genetic) correlation coefficients were -0.375 (-0.987), -0.505 (-0.670) and -0.474 (-0.400) for these traits, respectively. FL had a significant positive correlation (P<0.05) with H. In addition, FL had significant positive correlations (P<0.01) with DBH and V. The phenotypic (genetic) correlation coefficients were 0.169 (0.980), 0.361 (0.828) and 0.328 (0.980) for these traits, respectively.
Generally, FL and growth traits showed significant positive correlations, while BWD and growth traits showed significant negative correlations. Neither MA nor HC had a significant correlation with other traits. Thus, MA and HC can be independently selected.
Multitrait index selection
The multi-trait selection index is a comprehensive index based on heritability and genotypic and phenotypic trait correlations which combines multiple target traits to obtain the greatest improvement. According to the ANOVA results, DBH, V, BWD, FWT, VP and FP (six traits in total) were significantly different among clones. Since MA and FL are usually used for the purpose of breeding, these and five other traits were selected as quantitative traits for further analyses. Eight traits (DBH, V, BWD, FL, MA, FWT, VP, FP) were used to build the index selection equation for comprehensive selection of growth traits and wood properties. Economic weight was estimated by the equal weight method; that is, the reciprocal of the standard deviation of each trait phenotype was used as the weight of the trait. These included DBH (x1), V (x2), BWD (x3), FL (x4), MA (x5), FWT (x6), VP (x7) and FP (x8). Economic weight vectors (W) were 0.7323, 74.1374, 47.6404, 0.0132, -0.3871, 1.6698, -0.2609 and 0.2739 for these traits, respectively. To reduce potential negative effects of traits in the exponential equation, the Kempthorne constraint index method was used to constrain the genetic progress of MA and VP to zero in order to maximize the genetic gain of the other traits.
The restricted and unrestricted selection index equations and the selection progress of trait breeding and are shown in Tab. 5. The partial regression coefficients of FWT and FP in equations I1, I2, I3, I4, I9, I10, I11 and I12 were negative, and the partial regression coefficients of MA and VP in equations I5, I6, I7 and I8 were negative, so these equations were not ideal. The selection progress and partial regression coefficients of all traits (including DBH, BWD and FL, but not MA) were positive in equation I14. The selection progress of comprehensive breeding was 0.8641, the estimated accuracy of comprehensive breeding was 0.7200, index heritability was 0.5520 and the selection progress of DBH, BWD, FL and MA was 0.4517, 0.0030, 29.6198 and 0.0000, respectively. Therefore, equation I14 was the best. According to the selection index equation, calculations of the index value of each clone are shown in Tab. 6.
Tab. 5 - The restricted and unrestricted selection index equations and the selection progress of trait breeding. (SP): the selection progress of comprehensive breeding; (Acc): estimated accuracy of comprehensive breeding; (Hi): heritability index. For a full description of predictor traits (x1, …, x8) , see the text.
| Kind | Selection index equations | SP | Acc | Hi |
|---|---|---|---|---|
| Unrestricted | I1=0.3802x1+30.9886x2+50.7049x3+0.0204x4+0.2625x5-0.3042x6+0.6773x7+0.8956x8 | 2.2270 | 0.9537 | 0.7730 |
| I2=0.5192x1+17.9449x2+47.8234x3+0.0165x4+0.1301x5-0.5377x6-0.0213x7 | 1.6626 | 0.8769 | 0.7093 | |
| I3=0.5657x1+4.5943x2+26.5315x3+0.1085x5-0.0582x6+0.0561x8 | 0.8634 | 0.7366 | 0.6125 | |
| I4=0.4813x1+19.4210x2+41.4966x3+0.0130x4-0.0228x5-0.3527x6 | 1.3683 | 0.7812 | 0.6985 | |
| I5=0.3272x1+57.6935x2+37.1581x3+0.0116x4+0.0873x5+0.0893x8 | 1.6657 | 0.8349 | 0.7326 | |
| I6=57.1845x2+48.1969x3+0.0101x4+0.0701x5+0.0889x8 | 1.3447 | 0.8920 | 0.8820 | |
| Restricted | I7=0.6471x1+29.2781x2+53.9793x3+0.0065x4+0.0216x5+0.6907x6+0.5655x7+0.4685x8 | 1.3860 | 0.5935 | 0.6467 |
| I8=0.6471x1+23.5232x2+52.7317x3+0.0073x4+0.0184x5+0.3134x6+0.1248x7 | 1.3317 | 0.7023 | 0.6368 | |
| I9=0.6807x1-8.3991x2+20.5807x3+0.0017x5-0.2080x6-0.0485x8 | 0.6885 | 0.5874 | 0.5572 | |
| I10=0.4505x1+16.3953x2+35.5167x3+0.0132x4-0.1088x5-0.4656x6 | 1.3436 | 0.7671 | 0.6479 | |
| I11=0.4656x1+39.3867x2+29.1130x3+0.0123x4-0.0454x5-0.0478x8 | 1.5264 | 0.7651 | 0.6084 | |
| I12=49.7285x2+38.9926x3+0.0109x4-0.0651x5-0.0457x8 | 1.1636 | 0.7719 | 0.7008 | |
| I13=0.1775x1+29.4083x3+0.0091x4-0.1150x5+0.3890x6 | 0.9723 | 0.6907 | 0.5230 | |
| I14=0.4318x1+27.5706x3+0.0067x4-0.1041x5 | 0.8641 | 0.7200 | 0.5520 |
Tab. 6 - Comprehensive evaluation of clones. (I): Selection index value.
| Clone | I | Clone | I | Clone | I |
|---|---|---|---|---|---|
| J19 | 16.75 | H7 | 15.51 | B5 | 15.08 |
| I18 | 16.29 | M19 | 15.50 | H4 | 15.08 |
| H16 | 16.24 | M14 | 15.47 | D13 | 15.02 |
| C13 | 16.14 | M10 | 15.46 | B8 | 14.94 |
| D23 | 16.10 | I5 | 15.43 | D11 | 14.90 |
| E9 | 15.92 | I10 | 15.40 | D22 | 14.82 |
| C9 | 15.89 | M20 | 15.32 | H8 | 14.80 |
| H13 | 15.89 | D16 | 15.26 | M16 | 14.77 |
| I8 | 15.87 | M15 | 15.24 | B6 | 14.75 |
| C16 | 15.86 | D17 | 15.19 | D5 | 14.49 |
| C12 | 15.82 | J20 | 15.17 | F6 | 14.44 |
| E8 | 15.81 | M13 | 15.13 | H12 | 14.39 |
| D24 | 15.79 | C15 | 15.11 | C14 | 14.34 |
| C1 | 15.70 | H5 | 15.10 | J9 | 14.27 |
| M17 | 15.60 | D9 | 15.09 | E5 | 13.89 |
Outstanding clone selectivity was 10 %, selected from the top four excellent clones (J19, I18, H16, C13). DBH, V, BWD, FL, MA, FWT, VP and FP of excellent clones were higher than all clones by 7.29%, 13.92%, 4.37%, 7.36%, -3.51%, 6.73%, -6.54% and 2.14 %, respectively.
Discussion
Variation and heritability
Numerous studies reported a wide range of genetic variation in growth and wood traits at the clonal level for forest tree species ([2], [31]). In this study, we found significant differences (P<0.05) in DBH, V, BWD, FWT, VP and FP among 45 clones of Pinus ussuriensis. This indicates that clonal effects in the joint analysis for growth traits and wood properties were significant.
Mean values for H, DBH and V of all clones ranged from 7.77 m to 12.44 m, 6.58 cm to 10.76 cm and 0.0175 m3 to 0.0537 m3, respectively. The GCVs of growth traits of all clones ranged from 2.70% to 14.61%. Among them, V had the largest GCV. Higher GCV means that clones showed a greater potential for improving traits. H2 of growth traits ranged from 0.084 to 0.663. This indicated that V and DBH were under moderate genetic control, while H was genetically controlled to only a mild degree.
The BWD mean values ranged from 0.2421 g cm-3 to 0.3083 g cm-3, smaller compared to previous studies of Populus tremuloides ([40]) and Populus tremula ([19]) clones. The broad-sense heritability of BWD was 0.499, higher than in previous studies ([42]). These differences might be due to different sampling methods, including different sampling heights and different cambial age of samples. FL mean values ranged from 677.01 µm to 855.56 µm, longer than those of P. ussuriensis ([42]) and Populus balsamifera L. ([17]) clones previously studied. The mean FL/W ranged from 26.96 to 35.04. The mean MA ranged from 9.30° to 18.67°. Density was generally considered one of the most important factors affecting wood properties such as stiffness, strength and shrinkage behavior ([12]). However, it has been shown both experimentally ([5]) and theoretically ([7]) that MA is at least as important as density for the prediction of strength and stiffness of solid wood. The results of this study showed smaller MA values than those reported for Betula pendula Roth ([4]). Decreased MA increases stiffness of cell walls ([6]). The mean CWP range was 32.13 to 46.87, similar to that reported in previous studies on poplars. Mean FWT ranged from 3.39 µm to 5.06 µm. In general, cell wall thickness is related to wood hardness, with greater thickness of the cell wall leading to greater hardness of wood. Mean VP, FP and RP ranged from 20.35% to 32.38%, 63.57% to 75.45% and 3.45% to 5.45%, respectively. Angiosperm wood consists mainly of axially elongated vessel elements and fibers, with radially elongated ray cells ([24]). Tissue proportion of different wood elements showed that wood fiber proportion is at maximum. Compared with the genus Quercus ([32]), the results of this study showed a higher fiber proportion and lower wood ray proportion. Mean HC ranged from 70.05% to 80.11%. HC showed a significant effect on the nature and utilization of the wood. The result was similar to previous studies of Populus deltoides Bartr. ([20]). From the perspective of PCVs, HC had the smallest and MA the largest values, while as for GCVs, HC had the smallest and VP the largest values. Broad-sense heritabilities ranged from 0.220 to 0.749, i.e., moderate to low values for wood properties. This is similar to previous studies by Atwood et al. ([1]) and Doran et al. ([11]). Combined with a modest coefficient of variation, genetic improvement through recurrent selection and breeding is promising.
Populus species are present across a broad range of climatic and edaphic conditions, bearing an important ecophysiological variability that often underlies inter and intraspecific adaptation patterns ([10]). Zhang et al. ([43]) have proven P. ussuriensis not only adapts to flat land and river beach but also to hillside land, with no disease or frost damage, thus having great growth potential ([43]).
Relationship between growth traits and wood properties
Sampling of wood properties is not only destructive and costly but also complicated. If the correlations among traits are already known, money and time can be saved by predicting or indirectly selecting unknown traits using known traits. Knowledge of the phenotypic and genotypic relationships between growth traits and wood properties is critical for their simultaneous genetic improvement.
Growth traits were all highly positive correlated. Growth traits and BWD were extremely negatively correlated, similar to Picea abies ([15]). This indicates the rapid growth rate was not conducive to the formation of large-density wood. There was significant positive correlation among DBH, FL and FP, which shows that diameter growth was beneficial to long fiber formation. In addition, DBH had significant negative correlation with VP, but the correlation coefficient was small. These results were similar to those found in Larix kaempferi ([27]). Moreover, we found no significant correlations between growth traits and FL/W, MA, CWP, FWT, RP or HC, indicating that these traits were independently inherited and could potentially be selected independently. On the other hand, we found negative correlations between growth and wood traits (BWD, FL/W, MA, CWP, VP and FP), similar to what was previously reported in poplar ([16]).
Multi-trait index selection
It is often difficult to comprehensively improve the yield and quality of trees due to negative correlations between growth and wood traits. The selection index is a helpful tool for guiding this process, allowing for multiple features of interest to be selected at one time ([26]). Christophe & Birot ([8]) and Nebgen & Lowe ([29]) used the multi-trait selection index to screen many provenances, clones and pedigrees for growth traits and wood properties.Guan et al. ([14]) used the multi-trait selection index to select superior clones of Populus deltoids × P. euramericana based on growth trait data, including wood properties and stem forms, of 33 12-year-old F1 clones. Aggregate character was improved with selection index, but the genetic gain of single characters using the selection index was lower than that obtainable from single-character selection. Zhou et al. ([46]) suggests that when using a selection index for joint selection of growth and material properties, the number of traits considered in constructing the selection index equations should not be excessive. In this study, eight quantitative traits were selected as predictors according to the results of ANOVA, with correlations between traits and breeding objectives. The best equation was chosen according to breeding objectives, selection progress and estimated accuracy of comprehensive breeding. Based on the selection index equation and selectivity (10%), the top four clones (J19, I18, H16, C13) resulted as superior clones. The averages of BWD, FL, FL/W, MA, CWP, FWT, VP, FP, RP and HC of these excellent clones were 0.2823 g cm-3, 824.13 µm, 31.04, 12.18°, 39.07%, 4.22 µm, 24.29%, 71.06%, 4.66%, 75.44%, respectively. The fiber length was 824.13 µm, meeting the slightly shorter fiber standard (700-900µm) specified by International Association of Wood Anatomists. The fiber length-width ratio satisfies the pulping performance standard (>30). The microfibril angle is 12.18°, the fiber strength increases with the decrease of microfibril angle. The fiber ratio and the holocellulose content were greater than 70%, which was suitable for pulp. Therefore, the selected clones can meet the requirements of the pulp and paper industry, being an excellent pulp material variety. Multi-trait index selection will be beneficial for future breeding programs, in particular when there is negative correlation between growth traits and wood properties.
Conclusion
This study assessed the genetic variation of growth traits and wood properties of 45 P. ussuriensis clones. Significant differences between clones for most growth traits and wood properties reflected the selective breeding value of clones. Analysis of broad-sense heritability showed that growth traits and wood properties are genetically controlled only at moderate to low levels. We found negative correlations between growth traits and most wood properties. To improve growth and wood traits simultaneously, the multi-trait selection index method was used to pick out the excellent clones. Combined with the survival rates of clones, the three clones selected (I18, H16, C13) showed higher values than all other clones by 2.83%, 9.81%, 3.40%, 6.59%, -7.54%, -0.39%, -2.12% and 0.31% for diameter at breast height, volume, basic wood density, fiber length, microfibril angle, fiber double wall thickness, vessel proportion and wood fiber proportion measurements, respectively. The results demonstrated that this method is feasible, and combined selection for growth and wood properties is effective for tree improvement.
The conclusions of this study is based on a single site, but it could be used for the further studies on breeding and genetic improvement of P. ussuriensis. In addition, the study on microfibril angle, cell wall percentage, fiber double wall thickness and tissue proportion is conducive to the selection of high-quality pulpwood clones.
Acknowledgements
Jiaojiao Jin and Kailong Li conceived and designed the experiments. Jiaojiao Jin, Xiyang Zhao, Huanzhen Liu, Ziwen Song and Xuyu Ma performed the experiments. Kailong Li provided reagents, materials and tools and provided careful guidance. Jiaojiao Jin analyzed the data, summarized the results and wrote the manuscript. Kailong Li, Xiyang Zhao, Huanzhen Liu and Sui Wang revised the manuscript.
This work was supported by the National Key R & D Program of China (Grant No. 2016YFD0600404).
References
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Xiyang Zhao
Huanzhen Liu
Sui Wang
Ziwen Song
Xiaoyu Ma
Kailong Li
State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, 150040 (China)
Corresponding author
Paper Info
Citation
Jin J, Zhao X, Liu H, Wang S, Song Z, Ma X, Li K (2019). Preliminary study on genetic variation of growth traits and wood properties and superior clones selection of Populus ussuriensis Kom.. iForest 12: 459-466. - doi: 10.3832/ifor2991-012
Academic Editor
Fulvio Ducci
Paper history
Received: Nov 14, 2018
Accepted: Jul 18, 2019
First online: Sep 29, 2019
Publication Date: Oct 31, 2019
Publication Time: 2.43 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2019
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 42983
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 36117
Abstract Page Views: 3195
PDF Downloads: 2848
Citation/Reference Downloads: 2
XML Downloads: 821
Web Metrics
Days since publication: 2318
Overall contacts: 42983
Avg. contacts per week: 129.80
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2019): 5
Average cites per year: 0.71
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Age trends in genetic parameters for growth and quality traits in Abies alba
vol. 9, pp. 954-959 (online: 07 July 2016)
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
A comparative study of growth and leaf trait variation in twenty Cornus wilsoniana W. families in southeastern China
vol. 10, pp. 759-765 (online: 02 September 2017)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Stability analysis and selection of optimal Eucalyptus urophylla × E. grandis families
vol. 18, pp. 293-300 (online: 20 October 2025)
Research Articles
Comparison of range-wide chloroplast microsatellite and needle trait variation patterns in Pinus mugo Turra (dwarf mountain pine)
vol. 10, pp. 250-258 (online: 11 February 2017)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Comparison of genetic parameters between optimal and marginal populations of oriental sweet gum on adaptive traits
vol. 11, pp. 510-516 (online: 18 July 2018)
Review Papers
Genetic diversity and forest reproductive material - from seed source selection to planting
vol. 9, pp. 801-812 (online: 13 June 2016)
Research Articles
Genetic variation of Fraxinus excelsior half-sib families in response to ash dieback disease following simulated spring frost and summer drought treatments
vol. 9, pp. 12-22 (online: 08 September 2015)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword