Total RNA extraction from strawberry tree (Arbutus unedo) and several other woody-plants
iForest - Biogeosciences and Forestry, Volume 1, Issue 4, Pages 122-125 (2008)
doi: https://doi.org/10.3832/ifor0465-0010122
Published: Aug 27, 2008 - Copyright © 2008 SISEF
Research Articles
Abstract
Studies of plant gene expression today need pure preparations of high-yielding undegraded RNA. This is not easily accomplished when working with plants and tissues like strawberry tree (Arbutus unedo) leaves that accumulate large amounts of polysaccharides and polyphenolic compounds, which co-purify with RNA. An improved leaf-tissue protocol developed for gene expression studies on Arbutus unedo yields for the first time a purity of RNA extract that makes possible cDNA synthesis and qPCR analysis in this plant species. When tested on material considered recalcitrant (leaves, roots, fruit flesh, fruit peel and styles from Pyrus communis, Prunus avium, Prunus persica and Cydonia oblonga), the method was able to extract RNA with good yield and high purity. This scalable, phenol-free, fast and easy-to-use RNA extraction protocol is effective on Arbutus unedo leaves as well as on awide range of different species and tissues, thus resulting particularly useful for gene expression analysis in non-model species for molecular biology.
Keywords
Arbutus unedo, RNA extraction, Roots, Styles, Woody plants, Fruits
Introduction
RNA extraction from specific plant organs and tissues is a preliminary step for many molecular studies in plant biology. However, isolation of high-quality RNA from some tissues is often difficult due to the high amounts of polyphenols, polysaccharides and other secondary metabolites they accumulate. These contaminants tend to co-purify and co-precipitate with the RNA in the presence of alcohols, thereby remaining in the final extracts and interfering with such downstream applications as cDNA synthesis, restriction endonuclease enzyme digestion and the establishment of cDNA libraries ([16]). These problems often occur during RNA extraction from recalcitrant plants and, especially, from reserve-rich organs like fruits and seeds. In addition, the biosynthesis and accumulation of secondary metabolites in plants under biotic and abiotic stress, such as pathogen infection and drought, can be significantly enhanced ([22], [1]). While kits supplied by biotechnology companies extract RNA successfully from many tissues, they proved ineffective on tissues rich in polyphenols or polysaccharides ([8]).
Several protocols for RNA isolation from tissues of species with high contents of polyphenols or polysaccharides have been reported, including methods using soluble polyvinylpyrrolidone and ethanol precipitation ([16]), hot borate ([20]), phenol extraction ([9]), calcium precipitation ([3]), 2-butoxyethanol ([13], [14]) or cetyl trimethyl ammonium bromide (CTAB) in the extraction buffer ([15]). We tested four of these protocols ([9], [13], [14], [15]) and two commercially available RNA extraction kits to secure high-quality RNA in good amounts from an extremely recalcitrant plant such as strawberry tree (Arbutus unedo); the overall results proved unsatisfactory for all the methods, although the one described by Meisel et al. gave better results in terms of RNA purity compared with the others. We thus developed and tested a modified protocol that, through a simple optimization of some critical steps, allowed to achieve a great improvement in both RNA yield and purity when extracting from strawberry tree leaves. The effectiveness and versatility of this method was subsequently tested on different tissues (styles, fruit peel, fruit flesh, roots, leaves and seeds) from Pyrus communis, Prunus avium, Prunus persica and Cydonia oblonga, yielding in all cases adequate RNA amounts with a good purity grade.
Materials and methods
Plant material
All plant materials used for RNA extraction were grown in the experimental fields of the Department of Fruit Tree and Woody Plant Sciences (Cadriano, Bologna, Italy). The tissues were completely differentiated and healthy. Pear tree styles were harvested immediately before bloom and fruit peel collected in the course of ripening. Year-old leaves were gathered from strawberry trees (evergreen) and the finest roots were collected from quince tree. Cherries were processed when commercially ripe for both seeds and fruit flesh, and adult leaves were collected from Peach.
Solutions
All solutions and reagents were prepared in diethylpyrocarbonate (DEPC)-treated water ([17]) and autoclaved. Tris-HCl (pH 8.0) was added to the appropriate solutions after autoclaving.
- Extraction buffer: 2% (w/v) CTAB, 2% (w/v) PVP (mol wt 40.000), 100 mM Tris-HCl (pH 8.0), 25 mM EDTA, 2 M NaCl, 0.05% spermidine trihydrochloride (w/v), 2% ß-mercaptoethanol (added just before use).
- Resuspension buffer: 1 M NaCl, 0.5% SDS, 10 mM Tris HCL (pH 8.0), 1 mM EDTA (pH 8.0)
- Chloroform:isoamyl alcohol (24:1 v:v)
- 8 M LiCl
- Absolute Ethanol
RNA extraction protocol
Extraction buffer (1 ml per 100 mg of Arbutus unedo leaves; see Tab. 2 for quantities used when extracting from other tissues) was pre-heated to 65°C in a water bath. Leaves were ground in liquid nitrogen by pestle and mortar and the frozen powder quickly transferred to the extraction buffer. The sample was mixed thoroughly and incubated at 65 °C for 15 min, during the incubation the tube was vortexed 8-10 times to prevent the separation of tissue debris from extraction buffer.
Tab. 2 - Yield and purity of RNA extracted from different tissues. RNA extraction efficiency of the described protocol with tissues of several plant species. The weights of fresh tissue used for the extraction were around 20 mg for styles, 1 g for fruit peels and for fruit flesh, 500 mg for roots and for leaves and 50 mg for seeds. (a) Data represent mean and SE of 21 biological replicates; (b) Data represent mean and SE of 8 biological replicates; (c) Data represent mean and SE of 3 biological replicates.
| Species | Tissue | RNA yield | RNA purity | RNA purity |
|---|---|---|---|---|
| (µg/g FW) | A260/A280 | A260/A230 | ||
| Pyrus communis a | Style | 268 ± 40 | 2.06 ± 0.01 | 2.25 ± 0.03 |
| Pyrus communis b | Fruit peel | 7 ± 1 | 2.02 ± 0.03 | 1.9 ± 0.1 |
| Prunus avium c | Fruit flesh | 4.8 ± 0.7 | 2.09 ± 0.02 | 1.7 ± 0.1 |
| Cydonia oblonga c | Root | 40 ± 5 | 2.04 ± 0.02 | 1.41 ± 0.04 |
| Prunus persica c | Leaf | 68 ± 25 | 2.07 ± 0.06 | 2.07 ± 0.07 |
| Prunus avium c | Seed | 101 ± 14 | 2.12 ± 0.01 | 2.19 ± 0.01 |
An equal volume of chloroform:isoamyl alcohol (24:1 v:v) was added, the sample was vigorously vortexed and then centrifuged (15 min at 13000 g). The supernatant was transferred to a new tube and the extraction with chloroform:isoamylalcohol (24:1 v:v) repeated.
The supernatant was carefully transferred to a new tube without disturbing the organic phase and one-third volumes of 8 M LiCl was added. The sample was mixed thoroughly by inverting the tube and incubated overnight at 0 °C (in ice).
RNA was pelleted by centrifuging the sample at 15500 g for 35 min and resuspended in 500 μl of resuspension buffer. Samples were re-extracted with an equal volume of chloroform:isoamyl alcohol (24:1 v:v) to reduce residual contaminants; after centrifugation for 10 min at 15000 g the aqueous phase was recovered.
Two volumes of ice-cold 100% ethanol were added and RNA was precipitated at -80 ºC for 30 min. The sample was centrifuged at 17000 g for 20 min and the supernatant was removed. The pellet was air-dried at room temperature and then resuspended in 20-100 μl of DEPC-treated water.
Measurement of RNA yield and purity and downstream application
To prevent inaccurate quantification, the RNA pellet was slowly re-hydrated for 10 minutes on ice and mixed by inversion. The RNA yield was measured with a ND-1000 spectrophotometer (NanoDrop, Wilmington, Delaware USA) and the 260/280 and 260/230 absorption ratios were verified as quality indexes. The RNA was examined by electrophoresis on 1% agarose/TAE gels.
Total extracted RNA was treated with Turbo DNA-free (Ambion, Austin, USA) following the user’s manual and then a control PCR was carried out with 0.2 μg of total RNA in absence of reverse transcription in order to check for chromosomal DNA contamination. Samples that gave any amplification were purified a second time with DNase. Thereafter, 3 μg of pure RNA were used with M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, USA) and Oligo- (dT)20 or random primers for first strand cDNA synthesis following manufacturer’s instruction.
The first-strand cDNA obtained and the primers for 18S region (Forward: 5’-ACGGATCGCACGGCCTTCGTG-3’ and reverse; 5’-ACCAGACTTGCCCTCCAATGG-3’) were used to check the quality of the retrotranscription through PCR. The reaction was carried out in a MJ thermocycler using 20 μl of Platinum Blue PCR SuperMix (Invitrogen), 1 μl of reverse transcriptase mix and primers to a final concentration of 200 nM. Cycles were programmed as follows: one initial denaturing cycle at 94 °C for 3 min, followed by 30 cycles of 30 s of denaturing at 94 °C, 30 s of annealing at 55 °C, and 1 min elongation at 72 °C. The products were then loaded on agarose gel to verify amplification. Once verified, cDNA was used in a real time-PCR to quantify gene expression.
Results and Discussion
For successful isolation of pure, intact total RNA from Arbutus unedo, which is rich in polysaccharides compounds, it is important to prevent binding of these contaminants to nucleic acids. The success of an RNA isolation procedure thus depends on the quantity, quality and integrity of the recovered RNA ([19]). Four standard and improved methods for RNA isolation ([9], [13], [14], [15]) and two commercially available RNA extraction kits (RNeasy Plant MiniKitTM, Quiagen; SV Total RNA Isolation SystemTM, Promega), were applied to Arbutus unedo leaves. While the two kits recovered no RNA (Tab. 1), a sufficient amount of RNA was obtained with the others ([9], [13], [14]), but the purity was so low to make reverse transcription and successive gene expression analysis by real-time PCR impossible (Tab. 1).
Tab. 1 - Yield and purity of RNA extracted from Arbutus unedo by different protocols. Extraction results of existing protocols on strawberry leaves. The method described in this study is presented with an intermediate developing step (I) and with the final protocol described in the Materials and methods section (II). See text for details. (A) Data represent mean and SE of 8 biological replicates; (B) Data represent mean and SE of 28 biological replicates; (C) Data represent mean and SE of 6 biological replicates; (D) Data represent mean and SE of 16 biological replicates; (E) Data represent mean and SE of 58 biological replicates.
| Methods | RNA yield (µg/g FW) |
RNA Purity (A260/A280) |
RNA Purity (A260/A230) |
|---|---|---|---|
| RNeasy Plant Mini KitTM Quiagen | 0 | n.d. | n.d. |
| SV Total RNA Isolation SystemTM, Promega | 0 | n.d. | n.d. |
| Komjanc et al. ([9])A | 29 ± 7 | 0.8 ± 0.2 | 0.9 ± 0.2 |
| Manning ([14])A | 27 ± 12 | 1.6 ± 0.1 | 0.8 ± 0.1 |
| Malnoy et al. ([13])B | 49 ± 18 | 1.02 ± 0.04 | 0.5 ± 0.1 |
| Meisel et al. ([15])C | 3 ± 1 | 1.6 ± 0.1 | 1.9 ± 0.1 |
| Method described in this study ID | 18 ± 4 | 1.5 ± 0.1 | 1.9 ± 0.1 |
| Method described in this study IIE | 22 ± 2 | 1.86 ± 0.04 | 2.02 ± 0.01 |
Even the protocol reported by Meisel et al. ([15]) proved inadequate. Although it extracted good quality RNA probably thanks to the CTAB in the extraction buffer, which dampens polysaccharide noise ([5]), its yield was very poor, thereby precluding single-leaf gene expression analysis (Tab. 1). Accordingly, we modified Meisel’s protocol to improve RNA yield without losing the high quality of the extract. In fact, a technique improvement was essential to further molecular studies of a plant like strawberry tree.
Several critical steps that could affect the extraction yield and quality have been identified in the Meisel’s protocol when applied to strawberry tree leaves. The ratio of sample fresh weight to extraction buffer, in particular, seems to play a crucial role. The first yield improvement, which was from 3 to 18 μg RNA/g FW (see Tab. 1, “method described in this study I”) was in fact achieved decreasing the quantity of starting fresh tissue from 2.5 g to 1 g per 1ml of extraction buffer, and raising the centrifuge speed to ensure a better separation of debris from the aqueous, RNA-containing phase.
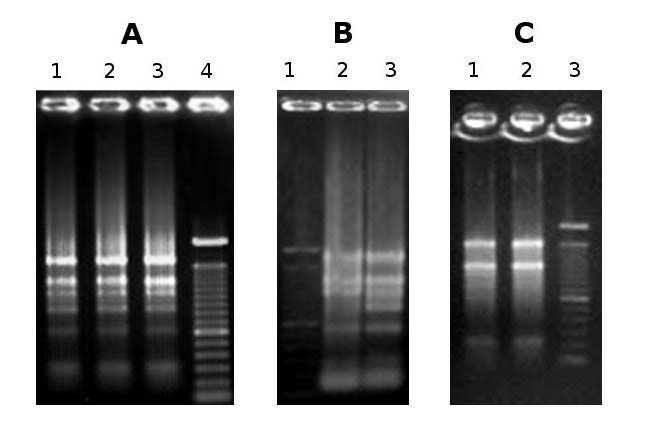
Subsequently, a further optimization of extraction was obtained by further reducing the sample fresh weight to the final value of 100 mg per 1 ml extraction buffer. Moreover, the lithium chloride (LiCl) precipitation step was carried out in ice instead of at 4°C; this seemed to assure a better precipitation efficiency and RNA integrity preservation. These changes resulted both in a better yield (up to 21 μg RNA/gFW) and RNA purity, determined by A260/280 and A260/230 absorption ratios (see Tab. 1, “method described in this study II”; the complete protocol is described in the “Materials and methods” section). The RNA resulted intact after agarose gel electrophoresis (Fig. 1) and suitable for reverse transcription (Fig. 2) and real time-PCR.
Fig. 1 - Agarose gel electrophoresis of RNA. Total RNA extracted from: (A) Cydonia oblonga root (lane 1), Prunus avium fruit flesh (lane 2) and seed (lane 3), (B) Prunus persica (lane 2) and Arbutus unedo (lane 3) leaves, (C) Pyrus communis style (lane 1) and fruit peel (lane 2). The 28s rRNA band appears equal to or more abundant than the 18s rRNA band, thereby indicating that little or no RNA degradation occurred during extraction.
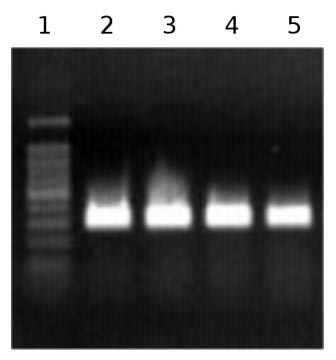
Fig. 2 - Agarose gel electrophoresis of PCR products. Partial 18S rRNA sequence amplification after retrotranscription from Arbutus unedo RNA. Lanes 2-5 are the products obtained from different RNA extracts.
As noted above, plant tissues ranging from roots to fruit flesh of various woody species were then tested for the quality and yield of the RNA extracted. Although our protocol was originally developed for the particularly difficult plant tissues of Arbutus unedo leaves, it proved notably versatile for other species and tissues, resulting in high-quality RNA in all cases (see Fig. 1 and Tab. 2). As mentioned above, the weight of starting fresh material per ml of extraction buffer is a critical parameter that had to be adjusted in order to succeed in extracting RNA from strawberry tree leaves. However, when extracting from the other tested species and tissues, the method proved to be effective with a wide range of starting material amounts, giving good results from as little as 20 mg of pear styles, and from 1 g of pear fruit peel or sweet cherry fruit flesh.
Conclusions
The small but significant changes we made to Meisel’s protocol ([15]) made possible to obtain a pure RNA extract from strawberry tree leaves while, to ourknowledge, no other studies report RNA extraction from this plant. The development described improved extraction yield and purity while cutting extraction time.
Compared to other protocols developed for RNA extraction from recalcitrant species and/or tissues ([11], [18], [6], [12], [2], [4], [10], [7], [21]), the presented method combines the advantages of being phenol-free, easily applicable to a wide range of tissues and not requiring expensive commercial kits.
Moreover, the varying amounts of starting material needed for different tissues (from 20 mg of styles to 1 g of fruit flesh or peel) enabled testing of the protocol’s scalability. Indeed, requiring only minor adjustments in labware and grinding because of material variability, our method also proved to be notably versatile and of good scalability, making it possibly the preferred “choice” for not expensive plant RNA extraction.
List of abbreviations
CTAB: cetyltrimethylammonium bromide; DEPC: diethylpyrocarbonate.
Acknowledgements
AZ performed the protocol elaboration, carried out the extractions from Arbutus unedo leaves and drafted the manuscript. LP carried out the extraction from Pyrus communis fruit peels, quince roots and cherry tree, helped to improve the protocol and participated in manuscript drafting. PDF carried out the extraction on Pyrus communis styles and participated to manuscript revision.
The authors thank Dr. G. Tonon for fruitful discussion and helpful assistance during protocol development, Dr. L. Dondini and Dr. S. Musacchi for helpful suggestions and practical support, Prof. Sansavini for encouragement and the availability of his lab and a special thanks to Anna for her strength.
References
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
L Pierantoni
P De Franceschi
Department of Fruit Tree and Woody Plant Science, University of Bologna, v. Fanin 42, Bologna (Italy)
Corresponding author
Paper Info
Citation
Zamboni A, Pierantoni L, De Franceschi P (2008). Total RNA extraction from strawberry tree (Arbutus unedo) and several other woody-plants. iForest 1: 122-125. - doi: 10.3832/ifor0465-0010122
Academic Editor
Marco Borghetti
Paper history
Received: Feb 18, 2008
Accepted: Jun 27, 2008
First online: Aug 27, 2008
Publication Date: Aug 27, 2008
Publication Time: 2.03 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2008
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 79794
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 66178
Abstract Page Views: 4717
PDF Downloads: 7636
Citation/Reference Downloads: 116
XML Downloads: 1147
Web Metrics
Days since publication: 6364
Overall contacts: 79794
Avg. contacts per week: 87.77
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2008): 36
Average cites per year: 2.00
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Genetic diversity of core vs. peripheral Norway spruce native populations at a local scale in Slovenia
vol. 11, pp. 104-110 (online: 31 January 2018)
Review Papers
Genetic diversity and forest reproductive material - from seed source selection to planting
vol. 9, pp. 801-812 (online: 13 June 2016)
Research Articles
Age trends in genetic parameters for growth and quality traits in Abies alba
vol. 9, pp. 954-959 (online: 07 July 2016)
Research Articles
Comparison of genetic parameters between optimal and marginal populations of oriental sweet gum on adaptive traits
vol. 11, pp. 510-516 (online: 18 July 2018)
Research Articles
Fine-scale spatial genetic structure in a multi-oak-species (Quercus spp.) forest
vol. 8, pp. 324-332 (online: 05 September 2014)
Research Articles
Genetic control of intra-annual height growth in 6-year-old Norway spruce progenies in Latvia
vol. 12, pp. 214-219 (online: 25 April 2019)
Research Articles
Testing a dual isotope model to track carbon and water gas exchanges in a Mediterranean forest
vol. 2, pp. 59-66 (online: 18 March 2009)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword