SimHyb: a simulation software for the study of the evolution of hybridizing populations. Application to Quercus ilex and Q. suber suggests hybridization could be underestimated
Álvaro Soto , David Rodríguez-Martínez, Unai López De Heredia
iForest - Biogeosciences and Forestry, Volume 11, Issue 1, Pages 99-103 (2018)
doi: https://doi.org/10.3832/ifor2569-011
Published: Jan 31, 2018 - Copyright © 2018 SISEF
Research Articles
Collection/Special Issue: INCOTW - Sassari, Italy (2017)
International Congress on Cork Oak Trees and Woodlands
Guest Editors: Piermaria Corona, Sandro Dettori
Abstract
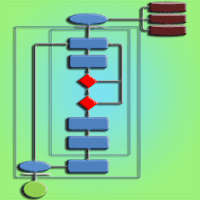
We present SimHyb, a Java-based software for the simulation of mixed hybridizing populations. The software incorporates user-defined initial parameters and input files to account for the initial census size of two species in a closed population, the number of intermediate specific classes, the directional fertility among specific classes, the fitness coefficients for each specific class, the inheritance of fitness, and the degree of ageing and self-incompatibility of the individuals. All these demographic and adaptive parameters can be modified by the user to analyze their effect on the evolution of the mixed population. SimHyb allows the traceability of each individual, whose pedigree is also recorded. For each simulated generation the software yields an output file that is easily convertible to an input for Structure, one of the most popular softwares for the Bayesian analysis of populations. Application of SimHyb to simulate Quercus ilex and Q. suber hybridizing populations, and further analysis with Structure, reveals that advanced introgressed individuals are very often misclassified with the currently available set of nuclear microsatellite markers, so that introgression between these two species could have been underestimated in previous studies. However, we provide a simple parameter based on Structure results to identify the directionality of pollination in the progeny of a known mother tree.
Keywords
Hybridization, Introgression, Simulations, Molecular Markers, Quercus suber, Quercus ilex
Authors’ Info
Authors’ address
David Rodríguez-Martínez
Unai López De Heredia
GI Genética, Fisiología e Historia Forestal, Dept. Sistemas y Recursos Naturales, ETSI Montes, Forestal y del Medio Natural, Universidad Politécnica de Madrid, Madrid (Spain)
Corresponding author
Paper Info
Citation
Soto Á, Rodríguez-Martínez D, López De Heredia U (2018). SimHyb: a simulation software for the study of the evolution of hybridizing populations. Application to Quercus ilex and Q. suber suggests hybridization could be underestimated. iForest 11: 99-103. - doi: 10.3832/ifor2569-011
Academic Editor
Piermaria Corona
Paper history
Received: Jul 28, 2017
Accepted: Jan 12, 2018
First online: Jan 31, 2018
Publication Date: Feb 28, 2018
Publication Time: 0.63 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2018
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 49513
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 42026
Abstract Page Views: 3129
PDF Downloads: 3118
Citation/Reference Downloads: 9
XML Downloads: 1231
Web Metrics
Days since publication: 2960
Overall contacts: 49513
Avg. contacts per week: 117.09
Citation Metrics
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2018): 10
Average cites per year: 1.25
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
References
Whole genome resequencing reveals diagnostic markers for investigating global migration and hybridization between minke whale species. BMC Genomics 18 (1): 83.
CrossRef | Gscholar