Response of soil bacterial communities to nitrogen and phosphorus additions in an age-sequence of subtropical forests
iForest - Biogeosciences and Forestry, Volume 14, Issue 1, Pages 71-79 (2021)
doi: https://doi.org/10.3832/ifor3655-013
Published: Feb 11, 2021 - Copyright © 2021 SISEF
Research Articles
Abstract
With global nitrogen (N) deposition continuously increasing, few reports exist describing how soil bacteria respond at the molecular level to long-term addition of excess N in variously aged forests. To reveal the responses of soil bacteria to the elevated N, an experiment was conducted with a chronic nutrient addition within differently aged stands (46-, 78-, and 200-years-old) in the northern subtropical China since 2011, including three treatments, CK (no N nor phosphorus (P) additions), N treatment (N, 100 kg N ha-1), and N with P (N+P, 100 kg N ha-1 + 50 kg P ha-1) to examine potential P limitation under N deposition. Metagenomic sequencing was used to examine the snapshot responses of soil bacterial communities. Soil moisture and texture, ammonium, nitrate, SOC (soil organic carbon), TN (soil total nitrogen), TP (total phosphorus), DOC (dissolved organic carbon), DON (dissolved organic nitrogen) were measured to explain the influence mechanism of forest age and fertilization on changes of microbial community. Following N addition, soil bacterial community diversity and most dominant phyla increased, but they showed a decrease with increasing stand age. The effects of fertilization on the same taxa were variable across forest ages. Soil bacterial community responded differently in 7-year fertilization, with distinct shift in 46-year-old forest and adaptability to long-term N addition in the 200-year-old forest. Soil texture and moisture, DOC, DON, pH, SOC/TN and TP were significantly correlated with bacterial community across stand ages, while N fertilization affected the bacterial community mostly via inducing soil moisture, NO3--N, DOC and pH in the 46-year-old forest, whose effects decreased with increasing stand age. Our results suggest that due to the variation of soil physicochemical properties among forest ages, soil bacterial communities are more stable and resilient to N deposition with increasing the age of stands. Soil bacterial communities might not encounter P limitation following the long-term addition of N in the subtropics.
Keywords
Nitrogen Deposition, Subtropical Evergreen Broadleaved Forest, Forest Age, Metagenomic, Soil Bacterial Community
Introduction
Nitrogen (N) is one of the key elements involved in ecosystem functioning. Currently, due to the use and emission of active N, China has become the third largest N deposition area after Europe and North America. Total nitrogen oxide (NOx) emissions increased from 8.4 Tg yr-1 in 1990 to 11.3 Tg yr-1 in 2000, whereas total NH3 emissions increased from 10.8 to 13.6 Tg during the same period ([23], [21]). Long-term inputs of inorganic N via atmospheric deposition may have a series of negative impacts on forest ecosystems, such as soil acidification ([38], [20], [46]), conspicuous changes in soil nutrient ([20]), and loss of plant community diversity ([51]).
Forest soils maintain the most diverse microbial habitats on earth, in which bacteria are the most abundant group of microorganisms ([13]). These bacteria contribute to a range of essential soil processes involved in the cycling of soil elements, and in mineral weathering that leads to the release of inorganic nutrients ([39]). The composition and functional group definition of bacterial phyla, genera and species have been reported in previous studies ([45], [26], [22]). The addition of N is considered to increase the availability of plant-derived C and/or N resources to meet the demands of microbial growth in the soils of most terrestrial ecosystems ([45], [26]). Additionally, changes in soil physicochemical properties resulted directly or indirectly from the addition of N could affect soil microbial communities ([29], [20]).
Due to bacteria exhibiting an extremely broad variety of metabolic types, the mechanisms underlying the microbial feedback of soils in response to global climate change remain elusive ([9], [22]). Nitrogen enters ecosystems largely through fixation, which is dominated by bacteria, accounting for more than 95% of the N input in undisturbed forests. The diversity of N-fixing bacteria was found to change rapidly with exogenous N inputs ([33]). Ramirez et al. ([32]) found that the abundance of Acidobacteria and Verrucomicrobia decreased, while Actinobacteria and Firmicutes increased, following the addition of N to a variety of soils. Apart from the modified composition of soil microbial communities, the biomass of soil bacteria reduced by up to 50% ([9]).
Several studies have revealed that bacterial diversity declines with the rate of N addition ([17], [42]), while a reduction in oligotrophic bacteria was often concomitantly found with decreased enzyme activity related to cellulose breakdown. This further supports that the N addition negatively affects microbial SOM (soil organic matter) decomposition. However, when the N addition rate was lower than 100 kg N ha-1 year-1 (about ten times higher than the normal global rate), a positive response on microbial growth was found ([55]). Distinct influences on the community composition of soil bacteria suggest that the effects of N deposition on soil microbes are likely ecosystem-specific ([22], [53]).
Significant changes in the composition of microbial communities have been reported during the development of forest stands ([15]). Among Eucalyptus plantations, the bacterial community diversity differed significantly with different ages, other than different vegetation species ([31]). Such shifts in bacterial community structures are largely driven by changes in resource availability and alteration of soil chemistry in response to forest succession ([20], [54]). Due to that the responses of bacteria to future high N deposition rates might be primarily contingent on bacterial composition, background acidification status or nutrient availability ([25], [54]), the responses of soil bacterial communities to N addition in differently aged forests may be significantly variable, and the initially younger sites with less-fertility may show greater compositional changes ([10]). However, few studies have been carried out to verify this hypothesis or specify which environmental factors exert the strongest effects on bacterial communities at different ages.
Although N and phosphorus (P) supplies are relatively closely balanced in most natural ecosystems ([7]), single element enrichment can lead to a limitation by the alternative nutrient. Soil microorganisms may also be limited by the availability of N or P ([40], [44]). As the result of high N deposition, P limitation may occur in soil bacterial communities. The addition of P could partially mitigate the negative effects of N addition on Shannon’s indices ([42]), as well as the biomass of soil bacteria ([5]).
Determining how relationships between site factors and bacterial communities affect the equilibrium between soil organic matter decomposition and C sequestration is vital for the prediction of responses of forest ecosystems to N deposition ([35]). Our study aimed to reveal the effects of N addition and N+P interactions on the richness and community composition of soil bacteria, as well as the underlying mechanisms in differently aged subtropical forests. We initially hypothesized that: (i) the heterogeneity of microbial community was distinct under different forest ages, meanwhile the effect of N-induced was partly offset by the change of environment with forest age, and the negative response of bacterial community in the older forest with stronger acidity and higher nutrient content to N deposition would be weaker; (ii) the addition of P to N fertilization would cause a relatively reduced decline in soil bacterial abundance, and lead to a change in soil bacterial community composition, by alleviating nutrient limitation for bacterial communities. Furthermore, based on the major physicochemical factors affecting soil bacterial community, we explored whether the mediators of microbial community variation were the same in different stands following a continuous seven-year nutrient additions.
Materials and methods
Study area and sites
The study area encompasses subtropical evergreen broadleaved forests of different ages in Dashan, Shitai County, Anhui Province in China (DS, 200-year-old - 30° 01′ 47″ N, 117° 21′ 23″ E), Zhawan Forest Farm, Qimen County (QM, 78-year-old - 29° 37′ 05″ N, 117° 32′ 30″ E), and Jitan, Shitai County (JT, 46-year-old - 30° 16′ 28″ N, 117° 26′ 23″ E) were selected (Fig. S1 in Supplementary material). The forest ages at the beginning of the experiment were determined based on the forest sub-compartment data of Forest Management Inventory (personal communication). The distance among sites is about 40 km, with an average elevation of 200-500 m a.s.l. This region belongs to the northern part of subtropical monsoon climate, and has distinct seasons (long winters and summers and a short spring and autumn). The annual average temperature is 16.0-16.6 °C, with the annual maximum of 38.8 °C and the minimum of -8.9 °C. The average annual precipitation ranges from 1521-1790 mm, of which 71% is primarily distributed during April-September. The experimental forests are secondary evergreen broad-leaved forest.
The dominant tree species were Castanopsis eyrei, C. sclerophylla, and Cyclobalanopsis glauca. The understorey vegetation was dominated by Camellia fraterna, Symplocos sumuntia, Lindera aggregate, and Woodwardia japonica. The detailed description of the experimental forests are given in Dai et al. ([6]) and Xu et al. ([47]). The soils of different plots are reddish yellow soils developed from the parent material of granite weathering. Moreover, to ensure the effects of the inherent soil properties (i.e., soil texture) and forest age, we conducted ribbon analysis using the procedure described by Thien ([37]) and sampled soil and conducted the fractionation analysis as described by Hunt & Gilkes ([14]). Both tests confirmed that soil texture of the all stands are sandy loam and silty loam (Tab. S1 in Supplementary material).
Experimental design
Based on the N deposition rate in the study area (bulk deposition 13 kg N ha-1 year-1), the treatments were designed as: CK (no N and P addition), N treatment (100 kg N ha-1 yr-1), and N+P treatment (100 kg N ha-1 yr-1 + 50 kg P ha-1 yr-1), with three replicate plots for each treatment. The experiment began in 2011 at the abovementioned three sites, with a completely randomized design.
The plot size was 20 × 20 m, with a 10-m wide buffer zone between adjacent plots. The N was applied in the form of NH4NO3, and P was in the form of Ca(H2PO4)2. The nutrients were dissolved in 20 L of stream water and evenly sprayed onto the corresponding plots near the soil surface using a backpack sprayer every two months from June 2011. The controls received the same amount of stream water (20 L) in each treatment event.
Sampling and measurements
In late July 2018, fresh soil samples from the 0-10 cm soil layers were extracted from each plot according to the five-point method, sieved (2 mm), thoroughly homogenized, and divided into two subsamples: one for soil analysis and another being stored at -80 °C for DNA extraction and subsequent molecular analysis. Soil bulk density (BD) was determined using a core sampler. The soil water content, ammonium (NH4+-N), nitrate (NO3--N), dissolved organic carbon (DOC), and dissolved organic nitrogen (DON) were measured with fresh soil.
Soil moisture was quantified by oven-drying at 105 °C to a constant weight. NH4+-N and NO3--N were measured by a FIAstar™ 5000 flow injection analyzer (FOSS Analytical, Hilleroed, Denmark). DOC and DON were extracted with a 0.5 M K2SO4 solution, and the extract was measured using a Multi 3100® C/N analyzer (Analytik Jena AG, Jena, Germany). The soil samples were air-dried for two weeks to a constant weight, and ground and passed though using a 0.154-mm sieve for measuring the soil organic carbon (SOC), total nitrogen (TN), pH, and total phosphorus (TP). SOC and TN were determined using an elemental analyzer (EA3000®, EuroVector, Pavia, Italy). After mixing water and soil to a ratio of 2.5:1, the pH of the soil solution was measured using a pH meter. Following wet digestion, the TP was quantified using a FIAstar™ 5000 flow injection analyzer. All measurements were repeated in triplicate.
Soil DNA extraction
The total microbial group DNA from different samples was extracted using an EZNA® Stool DNA Kit (D5625-02, Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer’s instructions. The total DNA was eluted in 50 μL of elution buffer and stored at -80 °C until measured by PCR from LC-Bio Technology (Hangzhou, China), with the isolation being confirmed by 1.2% agarose gel electrophoresis. After amplified by PCR, followed by purification, quantification, and library quality control. High throughput sequencing was performed with HiSeq4000® (Illumina Inc., San Diego, CA, USA) once the library was qualified. The sequencing mode was PE150.
Sequence analyses
Preprocessing of sequence data
Raw data was preprocessed by the software Cutadapt ver. 1.9 ([27]) for removing contained joints and low-quality data to obtain clean data for subsequent analysis. Subsequently, the sequence reads were scanned by the window method with 6 bp defaults. When the average quality value in the window was less than 20, the reads were cut off from window start to the 3′ end. Sequences of less than 60 bp in length, and sequences with an N content > 5% following truncation were abandoned. Finally, the host sequence contamination was removed ([16]).
Genome assembly
We initially assembled the genome of each sample using the software metaSPAdes ver. 3.10.0 ([1]). After a single sample was assembled, the unused reads of each sample were merged into a MIX sample for mixed assembly. The assembled result was a FASTA format file, in which a sequence (contig) was reserved for subsequent clustering analysis with contigs longer than 500 bp. Once the genome assembly was complete, we proceed to assemble the resulting evaluation. According to the assembly result, we employed the QUAST program matched with SPAdes to evaluate the result of the assembly ([4], [11]).
Gene prediction and abundance analysis
We used MetaGeneMark® for Linux 64bit (Gene Probe Inc., Atlanta, GA, USA) to predict the CDS (Coding Region) of each sample and mixed assembly contigs (≥ 500 bp), and filtered the sequence with CDS lengths of less than 100 nt using the software CD-HIT ver. 4.6.1 ([19]). Next, according to the CDS prediction results, non-redundant genes were obtained using CD-HIT software. Simultaneously, 95% identity and 90% coverage were used for clustering and the selection of the longest sequence as representative sequence. The software Bowtie2 ver. 2-2.2.0 ([18] - ⇒ http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) was used to align the clean data of each sample to gene sequences, and the number of reads of each gene of every sample was calculated. Unigenes for subsequent analysis were obtained once the number of reads ≤ 2 genes were filtered out and compared between the samples.
According to the number of reads and the length of each gene, the abundance information (TPM value) of each gene in each sample is calculated. The formula was as follows (eqn. 1):
where G represents the abundance of a unigene, K represents the K-th unigene, r is the number of reads for comparison of the above genes, and L is the length of the gene.
Species annotation
We compared the detected unigenes with the NR_mate Library to obtain species annotation information using the software Bowtie2. A rarefaction curve was developed to compare the species richness of the samples with different quantities of sequencing data, and to show the rationality of the amount of sequencing data. When the extraction sequence was 468.935, the rarefaction curves of different soil samples exhibited a tendency to flatten, which indicated that these sample sizes were sufficient to cover all bacteria, and the sequencing data covered most of the bacterial information in the sample (Fig. S2 in Supplementary material).
Data analysis
The alpha diversity indices included richness, Shannon-Weiner index (H′ - eqn. 2), and Simpson’s index (D - eqn. 3), and Chao1 index (eqn. 4), which were calculated at the species level. Richness refers to the sum of bacterial species in the soil. The diversity indices were calculated as follows:
where Ni is the number of individuals of the i-th species, N is the number of individuals of all species in the community, S is the overall number of species, and Pi is the proportion of individuals belonging to species i in all individuals; Sobs is the number of individuals of all species, F1 and F2 are the singleton and doubleton counts, respectively.
We tested the effects of the CK, N, and N+P treatments and forest age, and their interactions on the abundance of soil bacteria, alpha diversity (richness, Chao1, Shannon’s index, and Simpson’s index), and the abundance of groups with the relative abundances greater than 1% in the total bacterial community by performing two-way analysis of variance (ANOVA). We assessed the assumptions of normality for all models using the Shapiro test, histograms, and QQ normal plots of model residuals. We employed Levene’s test for the assumption of homogeneity. These analyses confirmed that both assumptions were met for all models.
To examine the effects of treatment and age on bacterial composition, we conducted a permutational multivariate analysis of variance (perMANOVA) implemented by the “adonis” function of the “vegan” package in R ([30]). In perMANOVA, we used a Bray-Curtis dissimilarity matrix to summarize species composition and employed 999 permutations to determine statistical significance. We visualized the compositional data using non-metric multidimensional scaling with a Bray-Curtis dissimilarity measure.
Further, we examined how the bacterial composition was linked to soil properties by using the function “envfit” of the “vegan” package ([30]). Soil parameters, which have significant effects (P<0.05) on bacteria as determined by the “envfit” function, were further used to examine the associations with soil bacterial abundance, alpha diversity and dominant phylum by Pearson’s correlation analysis. All analyses were performed in R ver. 3.6.0 ([34]).
Results
Effects of N and P addition on the abundance of bacterial communities
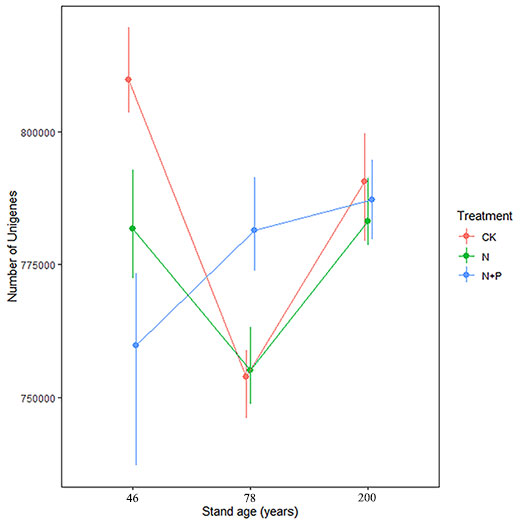
The effect of fertilization on the abundance of soil bacteria was strongly dependent on forest age (Tab. 1, Fig. 1). For CK, the bacterial abundance was lower in the 78-year-old stand than in the 46- and 200-year-old stands (Fig. 1). Compared with CK, both N and N+P treatments decreased bacterial abundance in the 46-year-old stand, with a greater reduction under the N+P treatment (Fig. 1). In the 78-year-old stand, the N+P treatment increased the bacterial abundance, while no effect appeared in the N treatment. However, in the 200-year-old stand, neither N nor N+P treatment apparently affected bacterial abundance.
Tab. 1 - Interactive effects of treatment and stand age on soil bacterial abundance, alpha diversity, and composition (n=9). P-values from ANOVA are displayed.
| Characteristic | Age | Treatment | Age × Treatment |
|---|---|---|---|
| Abundance | <0.001 | 0.076 | <0.001 |
| Richness | <0.001 | <0.001 | 0.001 |
| Chao1 | <0.001 | <0.001 | 0.111 |
| Shannon’s index | 0.148 | <0.001 | <0.001 |
| Simpson’s index | 0.110 | <0.001 | <0.001 |
Fig. 1 - Interactive effects of treatment and stand age on the abundance of soil bacteria (in terms of number of unigenes detected). Treatments are control (CK), nitrogen addition (N), and N and phosphorus addition (N+P). Values are bootstrapped means ± 95% confidence intervals (CIs) for individual samples and their sum (n=9). Differences between stand ages or treatments are significant (α = 0.05) when their CIs do not overlap.
Effects of N and P addition on alpha diversity of bacteria
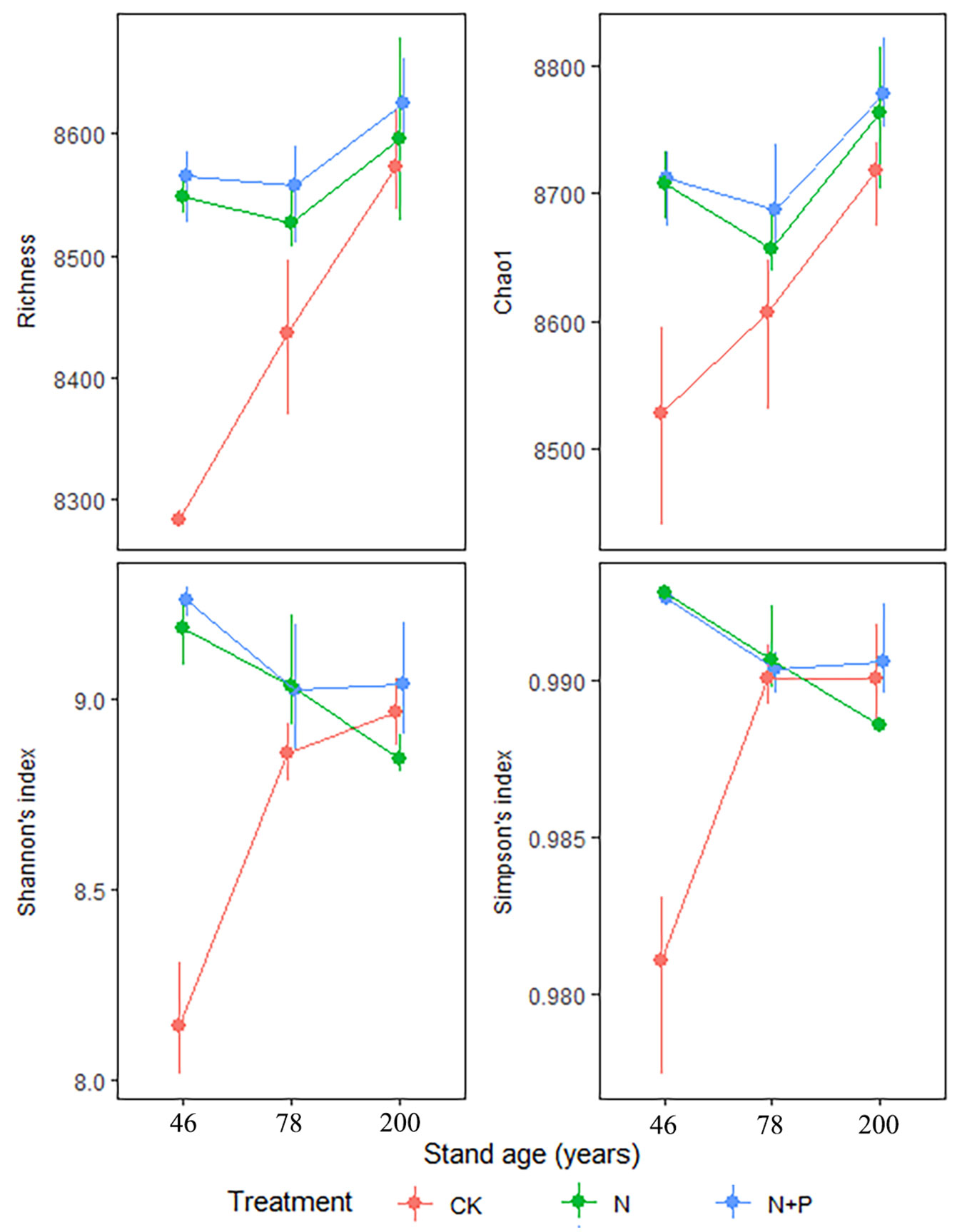
The species richness and Chao1 in CK increased with stand age, while Shannon’s and Simpson’s indices were lower in the 46-year-old stands compared with the older stands (Tab. 1, Fig. 2). Both N and N+P treatments increased species richness and Chao1 in the 46- and 78-year-old stands, with the greatest increases observed in the 46-year-old stand; however, increases in 200-year-old stands were far less pronounced. Similarly, both N and N+P treatments increased Shannon’s and Simpson’s indices in the 46-year-old stands but had little effect in the older stands (Tab. 1, Fig. 2).
Fig. 2 - Interactive effects of treatment and stand age on soil bacterial alpha diversity. Treatments are control (CK), nitrogen addition (N), and N and phosphorus addition (N+P). Values are bootstrapped means ± 95% confidence intervals (CIs) for individual samples and their sum (n=9). Differences between stand ages or treatments are significant (α = 0.05) when their CIs do not overlap.
Effects of N and P additions on the composition of bacterial communities
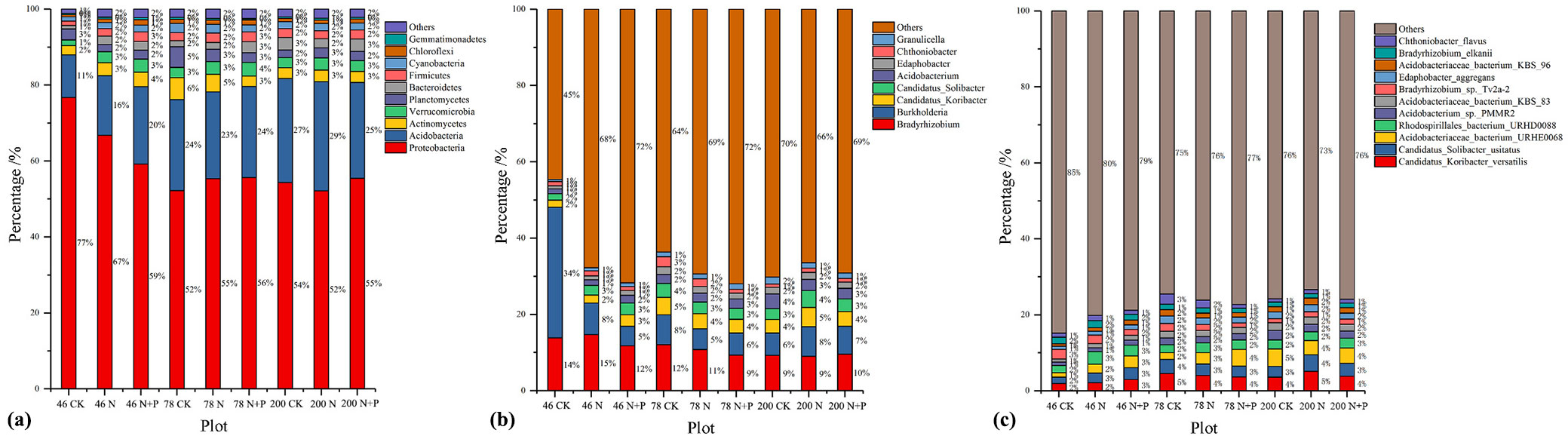
In different stands (Fig. 3a, Fig. 3b, Fig. 3c), the bacterial composition was dominated (>1%) by the phylum Proteobacteria (52-77%), followed by Acidobacteria (11-29%). The most frequent genera (>1%) for all stands were Bradyrhizobim (9-15%), Burkholderia (5-8%, except 6-34% in CK of the 46-year-old). The most prevalent bacterial species were Candidatus_Koribacter_versatilis (2-5%), and Candidatus_Solibacter_usitatus (2-4%).
Fig. 3 - Relative abundances of the soil bacterial groups (a: phylum level; b: genus level; c: species level) in each plot (n=9). Only groups with relative abundance > 1% are shown, while those with < 1% relative abundance are grouped into “others”.
The overall dominant bacterial composition, at the phylum, genus, and species levels, varied strongly with forest age and treatment (Tab. 2; Fig. S3, Fig. S4, Fig. S5 in Supplementary material). The abundance of Proteobacteria in 46-year-old stands was 58.4% and 46.5% higher than in 78- and 200-year-old stands. The phylum Verrucomicrobia, Cyanobacteria, Chloroflexi, Gemmatimonadetes, and genera Candidatus_Koribacter, Candidatus_Solibacter, Edaphobacter, Chthoniobacter in 78-year-old were higher than in the other two stands.
Tab. 2 - Interactive effects of treatment and stand age on soil bacterial phylum, genus, and species (n=9). P-values from ANOVA are displayed.
| Taxa | Age | Treatment | Age × Treatment |
|---|---|---|---|
| Phylum | <0.001 | 0.009 | <0.001 |
| Genus | <0.001 | <0.001 | <0.001 |
| Species | <0.001 | <0.001 | 0.111 |
In CK, the bacterial communities were more concentrated by the most abundant plyla and genera (e.g., Proteobacteria, Bradyrhizobyum, Burkholderia) in the 46-year-old stand than in the older stands. The responses of the same dominant bacterial communities to fertilization was contingent on forest age (Tab. S2, Tab. S3 in Supplementary material). It was observed that fertilization decreased Proteobacteria, and increased Actinomycetes, Verrucomicrobia, Firmicutes, Cyanobacteria, and Chloroflexi in the 46-year-old stands, while decreased Verrucomicrobia and increased Planctomycetes in the 78-year-old stands, and did not change at the phylum level in the 200-year-old stands.
At the genus level, fertilization increased Candidatus_Koribacter, Candidatus_Solibacter in 46- and 200-year-old stands, while both decreased in the 78-year-old. Moreover, fertilization increased Acidobacterium, Edaphobacter, Granulicella and decreased Burkholderia in the 46-year-old stands, reduced Bradyrhizobim, Burkholderia, Edaphobacter, and Chthoniobacter in 78-year-old stands, and decreased Acidobacterium and Granulicella in 200-year-old stands.
At the species level, fertilization increased Acidobacteriaceae_bacterium_URHE0068, Rhodospirillales_bacterium_URHD0088, and decreased Bradyrhizobium_sp_Tv2a2 in the 46- and 200-year-old stands. Moreover, Candidatus_Koribacter_versatilis and Candidatus_Solibacter_usitatus were reduced in 78-year-old stands but increased in 200-year-old stands by fertilization.
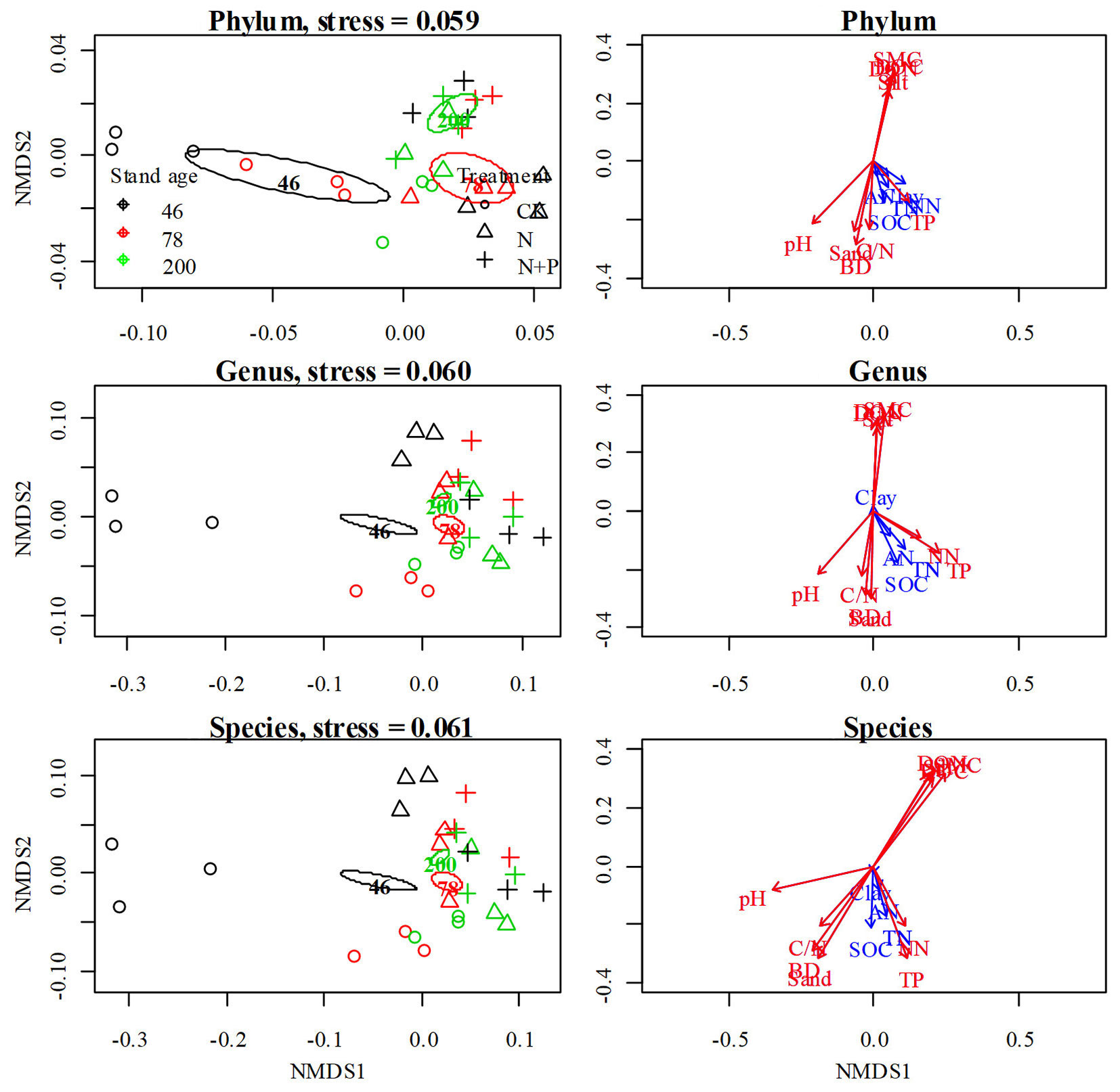
In CK, bacterial communities (phylum, genus, species level) formed independent groups by forest age, and the 46-year-old were grouped on the left and older sites on the right of axis 1 in the ordination space (Fig. 4a, Fig. 4b, Fig. 4c), which indicated that the younger stands possessed different overall species composition from the older stands. Moreover, three distinct fertilization type groups were prominently separated in the 46- and 78-year-old stands. For the same forest age, the degree of cluster separation formed by the treatments gradually decreased, which suggested the fertilization-induced effect was reduced with forest age.
Fig. 4 - Non-metric multidimensional scaling ordination of the composition of all bacterial phyla, genus and species of soil microbial communities. Close sites in the ordination space have similar bacterial assemblages, whereas those farther apart are less similar. Contours indicate time since reclamation. The length of vectors indicate the strength of associations; red vectors are significant (P<0.05).
Effects of soil physicochemical and vegetation factors on bacterial communities
The SOC/TN, pH, BD and soil sand content decreased, while the soil moisture, DOC, DON and soil silt content increased over time with more pronounced changes between stand ages (Fig. S6 in Supplementary material). Our result also indicated that changes of soil properties by fertilization varied between stand ages (Fig. S6 in Supplementary material). Further analysis revealed that in terms of bacterial species distribution, the interpretation rates of different physical and chemical factors were as follows: soil moisture (R2 = 0.70, P < 0.01) > DON (R2 = 0.61, P < 0.01) > DOC (R2 = 0.57, P < 0.01) > sand, silt (R2 = 0.58, P < 0.01) > pH, BD (R2 = 0.54, P < 0.001 - Tab. 3, Fig. 3d, Fig. 3e, Fig. 3f). Based on the different background, the effect of soil physiochemical properties on bacterial community changed among forest ages under fertilization. Correlation analysis (Tab. S2, Tab. S3 in Supplementary material) showed that soil moisture, NO3--N, DOC and pH were the main influencing factors in the 46-year-old, but these factors had little effect on 78- and 200-year-old stands among treatments.
Tab. 3 - Results of soil properties linked to the soil bacterial composition (at phylum, genus, and species level) using the function “envfit” in the vegan package (n=9). R2 and Pr(>r) represent the coefficient of determination and probability of significance, respectively.
| Variable | Phylum | Genus | Species | |||
|---|---|---|---|---|---|---|
| R2 | Pr (>r) | R2 | Pr (>r) | R2 | Pr (>r) | |
| Moisture content | 0.75 | 0.001 | 0.64 | 0.001 | 0.7 | 0.001 |
| BD | 0.59 | 0.001 | 0.51 | 0.002 | 0.54 | 0.001 |
| Sand | 0.42 | 0.004 | 0.54 | 0.001 | 0.58 | 0.001 |
| Silt | 0.43 | 0.002 | 0.5 | 0.001 | 0.58 | 0.001 |
| Clay | 0.04 | 0.633 | 0.01 | 0.98 | 0.01 | 0.97 |
| NH4+-N | 0.02 | 0.835 | 0.06 | 0.442 | 0.3 | 0.692 |
| NO3--N | 0.12 | 0.216 | 0.2 | 0.068 | 0.23 | 0.047 |
| DOC | 0.65 | 0.001 | 0.6 | 0.001 | 0.57 | 0.001 |
| DON | 0.6 | 0.001 | 0.58 | 0.001 | 0.61 | 0.001 |
| TP | 0.24 | 0.033 | 0.43 | 0.002 | 0.47 | 0.002 |
| TN | 0.07 | 0.432 | 0.17 | 0.99 | 0.13 | 0.192 |
| SOC | 0.15 | 0.168 | 0.24 | 0.046 | 0.19 | 0.083 |
| SOC/TN | 0.37 | 0.007 | 0.3 | 0.015 | 0.33 | 0.006 |
| pH | 0.63 | 0.001 | 0.51 | 0.001 | 0.54 | 0.001 |
Discussion
Molecular analyses provided insights into the fertilization effects on soil bacteria in forest ecosystems. In this study, the soil bacterial diversity in the CK showed an overall upward trend with forest age, which might be because older forests, due to the higher vegetation diversity, contain more plentiful available niches to promote the coexistence of more species of soil bacteria ([20], [31]). Many previous studies reported that the bacterial community diversity clearly declines with increasing N addition ([50], [17], [42]). In this study, such decline was only reflected by the Simpson’s and Shannon’s indices of the 200-year-old within the alpha index we considered, but this negative effect was relatively small (P > 0.05). Our result confirmed previous reports on the occurrence of different responses of soil bacterial abundance to fertilization between forest ages, with a significant decrease in young stands whilst the older stands had more stable bacterial communities ([9], [24], [46]).
The majority of the unigenes detected fell into four bacterial phylum categories. Proteobacteria constantly maintained the absolute dominance across different stand ages ([52]), followed by Acidobacteria and Actinobacteria, all of which are considered to be universally present in acidic soils ([43]). In our study, a decrease of Proteobacteria with increasing stand age was found, which was in contrast with previous studies reporting an increasing abundance of Proteobacteria along the succession ([15]). Previous studies revealed that soil bacteria of the same trophic type did not show uniformity subsequent to the nutrient addition for different vegetation types ([49], [29], [20]), which was further confirmed by our result. Although, a nutritional theory was proposed that the enhanced N availability increased the abundance of copiotrophic bacterial taxa, including Proteobacteria and Bacteroidetes, but lowered the proportion of oligotrophic Acidobacteria ([29]), which is further confirmed in the existing study of evergreen broad-leaved forests ([48]). However, the changes in bacterial species following fertilization in our study did not completely support the nutritional theory among forest ages, as we observed a decrease in the abundance of Proteobacteria (copiotrophic) in 46-year-old stands, and no significant change in older stands following N addition.
It was reported that fertilization results in a decline in diversity by stimulating the expansion of nitrophilous species and the competitive exclusion of others ([2], [29]). Compared with major phylum, the N-transformation-related phyla (e.g., Proteobacteria, Planctomycetes, Cyanobacteria) had no advantages as the result of N addition. The point was supported by several studies that the availability of NH4+-N rather than NO3--N was primarily responsible for the modification of bacterial diversity ([50], [29]). Although NH4+-N is the preferred nutrient for most bacteria ([28]), the change of NO3--N was larger than that of NH4+-N after fertilization in this study, which may also be the reason for the different results. Chen et al. ([3]) observed that soil total N and NO3--N were the main factors that influenced most variations of bacterial community composition in the agricultural ecosystems.
In this study, the effect of fertilization on soil bacterial community decreased with forest age. Bacterial community structure was altered by N treatment in 46-year-old stands, and the effects of N and N + P treatments differed significantly from CK. A weak effect of nutrient additions on soil bacterial diversity and community composition was found in 200-year-old stands in this study, which is similar to the results between primary and secondary tropical rainforests ([20]). Soil texture, nutrient availability and acidification may account for the variable response of soil bacteria to N addition ([25]). The differences in soil environmental regime between the different successional stages rather than chronic N fertilization might be the major control in soil bacterial composition in forest ecosystems. At the same time, the response of the dominant bacteria phyla to long-term N addition was distinct from that in short-term between forest ages ([5], [12], [36]). At our sites, the abundance of Proteobacteria was inhibited after 4-year N and N+P additions in both 46- and 200-year-old stands, while only occurred in the 46-year-old stands after 7-year N additions. This might reflect the acclimation to long-term high N deposition ([12]) and the elevated P availability with P addition, thus alleviating the disadvantages of chronic N enrichment ([42]).
Soil texture, moisture, NO3--N, DOC, and pH are important factors for predicting the distribution of soil bacterial composition, along with fertilization among forest ages ([41], [20], [54]). Our data revealed that fertilization-influenced soil bacteria mainly via change of soil moisture, NO3--N, DOC and pH in the 46-year-old stands, which had less effects in the older stands. Results from Wu et al. ([46]) confirmed a strongly negative correlation between soil bacterial alpha diversity, main phyla and pH. Furthermore, soil microbial communities were likely to be altered with the variations in soil moisture ([46]). The significant increase of soil moisture and decrease of soil pH with forest age might alleviate the effects to soil bacteria by fertilization ([42]). In the 46-year-old, the increase of bacterial diversity induced by soil moisture may be due to the decrease of the abundance of Proteobacteria and the increase of the abundance of other dominant flora, while the influence of pH is converse, but this regulation was not found in the older forest.
Background differences in soil C and N status between forest types have been thought to be possible causes for dissimilarity in soil bacterial community composition in tropical rainforests ([20]). Fertilization might indirectly affect soil microbes by modifying the soil C availability ([41]). When SOC/TN was >10, TN could be limited to soil microbes ([41]). In our sites, the SOC/TN was ranged from 12.32 to 15.38, so that TN might be a limiting nutrient to soil bacteria. Results from Wang et al. ([41]) showed that TP significantly affected soil bacterial communities, while our study showed that N+P addition did not cause pronounced changes in soil bacterial communities compared with N addition ([43]). The responses of soil microbes to P addition might vary with availability of other soil nutrients, as Esberg et al. ([8]) showed that microbial biomass and activity were more likely to be limited by P when C availability is high. Our results revealed that compared with N addition, the excessive P addition (N+P) exhibited negative effects for Proteobacteria in 46-year-old stands, as well as Cyanobacteria and Gemmatimonadetes, Verrucomicrobia, and Acidobacteria in 78-year-old stands. These results suggested that soil bacterial communities might not encounter P limitation following the long-term addition of P in N-rich forest ecosystems ([42], [43]).
Conclusions
The abundance of bacteria showed a concave distribution with forest age, while bacterial diversity showed an overall uptrend with ages in a chronosequence of subtropical forests. Soil bacterial community became more stable and resilient to nutrient additions with increasing of the stand age, with a significant decrease occurring only in the youngest stand within the age-sequence from 46- to 200-year-old. Moreover, the same soil bacterial group did not respond to nutrient addition consistently at different stand ages. Nitrogen transformation-related phylum had no advantages in N enrichment compared to others. Further, a prolonged N deposition can likely induce soil bacteria responses that are different from the short-term deposition. In this study, the driving factors of soil bacterial communities were forest age, followed by nutrient addition. The core bacterial communities were mainly controlled by soil texture, moisture, DOC, DON, pH, SOC/TN and TP among different stand ages.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (NSFC, No. 31770672 and 31370626) and the National Basic Research Program of China (2010CB950602). We gratefully acknowledge Professor. Han Y. H. Chen for his help in writing the manuscript. Thank the Forestry Bureau of Shitai County, and the Zhawan Nature Reserve, Anhui Province, for their providing fertilization experiment forests. We are thankful to Mr. Sun MY, Li PX, Wang F and Ma RH for sample collection and/or laboratory analysis. We also thank anonymous reviewers for comments which substantially improved this manuscript.
References
CrossRef | Gscholar
CrossRef | Gscholar
Gscholar
CrossRef | Gscholar
CrossRef | Gscholar
Gscholar
CrossRef | Gscholar
CrossRef | Gscholar
Authors’ Info
Authors’ Affiliation
Huiling Wang
Mengyuan Chen
Dandan Wang
Xiaoqin Cao 0000-0003-3894-1079
Bingyin Chu
Xiaoniu Xu 0000-0003-4126-2130
School of Forestry & Landscape Architecture, Anhui Agricultural University, Hefei 230036 (PR China)
Corresponding author
Paper Info
Citation
Dai Y, Wang H, Chen M, Wang D, Cao X, Chu B, Xu X (2021). Response of soil bacterial communities to nitrogen and phosphorus additions in an age-sequence of subtropical forests. iForest 14: 71-79. - doi: 10.3832/ifor3655-013
Academic Editor
Maurizio Ventura
Paper history
Received: Sep 17, 2020
Accepted: Dec 10, 2020
First online: Feb 11, 2021
Publication Date: Feb 28, 2021
Publication Time: 2.10 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2021
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 37864
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 31083
Abstract Page Views: 3418
PDF Downloads: 2748
Citation/Reference Downloads: 2
XML Downloads: 613
Web Metrics
Days since publication: 1821
Overall contacts: 37864
Avg. contacts per week: 145.55
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2021): 4
Average cites per year: 0.80
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Effect of exogenous nitrogen and phosphorus inputs on the microbe-soil interaction in the secondary Castanopsis sclerophylla forest in east China
vol. 11, pp. 794-801 (online: 14 December 2018)
Research Articles
Seasonal dynamics of soil respiration and nitrification in three subtropical plantations in southern China
vol. 9, pp. 813-821 (online: 29 May 2016)
Research Articles
Nitrogen deposition and its impact on forest ecosystems in the Czech Republic - change in soil chemistry and ground vegetation
vol. 10, pp. 48-54 (online: 29 June 2016)
Research Articles
Effects of arbuscular mycorrhizal fungi on microbial activity and nutrient release are sensitive to acid deposition during litter decomposition in a subtropical Cinnamomum camphora forest
vol. 16, pp. 314-324 (online: 13 November 2023)
Research Articles
The manipulation of aboveground litter input affects soil CO2 efflux in a subtropical liquidambar forest in China
vol. 12, pp. 181-186 (online: 10 April 2019)
Research Articles
Effects of tree species, stand age and land-use change on soil carbon and nitrogen stock rates in northwestern Turkey
vol. 9, pp. 165-170 (online: 18 June 2015)
Research Articles
The effect of clear-cut age on soil organic carbon and nitrogen indices in Scots pine (Pinus sylvestris L.) stands
vol. 18, pp. 146-153 (online: 09 June 2025)
Short Communications
Not all long-distance-exploration types of ectomycorrhizae are the same: differential accumulation of nitrogen and carbon in Scleroderma and Xerocomus in response to variations in soil fertility
vol. 14, pp. 48-52 (online: 18 January 2021)
Short Communications
Seasonal change in soil nitrogen mineralization in young Chamaecyparis obtusa stands at the upper and lower positions on a slope in central Japan
vol. 18, pp. 197-201 (online: 20 July 2025)
Research Articles
Fungal and bacterial communities in a forest relict of Pinus pseudostrobus var. coatepecensis
vol. 16, pp. 299-306 (online: 09 November 2023)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword