Effect of exogenous nitrogen and phosphorus inputs on the microbe-soil interaction in the secondary Castanopsis sclerophylla forest in east China
iForest - Biogeosciences and Forestry, Volume 11, Issue 6, Pages 794-801 (2018)
doi: https://doi.org/10.3832/ifor2673-011
Published: Dec 14, 2018 - Copyright © 2018 SISEF
Research Articles
Abstract
Soil microbes play a key role in the formation and decomposition of organic materials and in the improvement of the ecological environment. Despite continuous increases in nitrogen (N) and phosphorus (P) inputs due to the atmospheric deposition or fertilization, the response of soil to exogenous inorganic nutrients inputs remains elusive. By a manipulative experiment we simulated N and P depositions in a subtropical secondary Castanopsis sclerophylla forest in east China over the period 2011-2015, to evaluate the impact of inorganic nutrient addition on soil bacterial communities. Four treatments were administered (control and nutrient-enrichment: N, P, and N+P), with N added at a total of 100 kg N ha-1 yr-1 in the form of NH4NO3, and P at 50 kg P ha-1 yr-1 in Ca(H2PO4)2, sprayed near the soil surface at the end of each trimester. Quantitative PCR technique and Illumina platform-based sequencing analysis of the V3-V4 16S rRNA gene region were performed on total DNA extracted from soil samples to characterize the soil bacterial community abundance and diversity. As Proteobacteria, Acidobacteria and Actinobacteria were the predominant phyla in all conditions, treatments did not alter the distribution of bacterial phyla, while their relative abundances responded differently to N, P additions. A GLMM analysis showed that N input significantly (P < 0.05) reduced the relative abundance of Acidobacteria (13.11%), Bacteroidetes (0.6%), Elusimicrobia (0.21%), Nitrospirae (0.1%) and TM6 (0.04%). Relative abundance after P treatment significantly (P < 0.01) decreased for Nitrospirae (0.07%), and pronounced interactive effects of N and P additions (N:P) were observed on phylum Nitrospirae (P < 0.01) and TM6 (P < 0.05). Moreover, redundancy analysis revealed that soil pH was closely related to the bacterial community (r2 = 0.622, P = 0.015). Our findings suggest that exogenous N and P inputs affected the relative abundances and caused compositional shifts in the local bacterial community that closely associated with soil pH, thus providing the evidence that microbe-soil interactions are influenced by N and P availability in subtropical forest ecosystem of east China.
Keywords
16S rRNA, Nitrogen and Phosphorus Additions, Bacterial Populations, Soil PH, Subtropical Secondary Castanopsis sclerophylla Forest
Introduction
Atmospheric N deposition has increased dramatically since the industrial revolution due to fertilization and fossil fuel burning ([14], [42]), and the increases in N inputs greatly affect soil chemical properties ([34], [53]). A recent study suggests that N deposition has contributed to a 4.78% increase in the total carbon (C) storage of China’s forests between 1981 and 2010 ([17]). On the other side, N deposition causes soil acidification ([33], [53], [36]), which has become a significant threat to soil functions. He et al. ([19]) reported that the intensity of N deposition in China is equal or even exceeds those in Europe and America, raising general concerns from the government and the public. Some previous studies indicated that N deposition often stimulates forest growth and enhances carbon sequestration ([10], [50]), but the expected growth acceleration may be diminished when the accompanied P supply is deficient ([29]). In the context of low soil P contents in terrestrial ecosystems across most areas of China ([18]), the role of P deposition is relevant, as the high-level N deposition, which initially stimulated forest growth, is likely shifting large areas of China’s forests towards P limitation. Moreover, soil P availability in terrestrial ecosystems is primarily driven by mineral weathering, application of fertilizer inputs and atmospheric deposition ([54], [37]). Recent assessments of observed and modeled P deposition rates on a global scale ([35], [55]) showed a range of 0.01-1.0 kg P ha-1 yr-1 on land.
In forest ecosystems, dominant tree species influence the metabolic abilities of soil microbial communities directly through litter production and root exudation, and indirectly by affecting soil properties. Soil microbial communities interact with site-dependent factors controlling organic matter decomposition and carbon sequestration, providing a key to understand terrestrial ecosystem feedback to global climate ([46]). In general, N addition tends to decrease microbial diversity, increase the relative abundance of copiotrophic (i.e., fast-growing, low carbon-use efficiency) taxa (e.g., Proteobacteria and Bacteroidetes), and reduce that of oligotrophic (i.e., slow-growing, high carbon-use efficiency) taxa (e.g., Acidobacteria and Basidiomycota - [12], [27], [20]). On the other hand, soil bacteria constitute the main type of soil microorganisms, and play an extremely important role in soil structure and nutrient cycling ([40]), the formation and decomposition of organic matter ([2]), the regulation of plant growth ([15]), and the improvement of the ecological environment. Previous research has shown that bacterial community structure can reflect the soil current situation and tendency, indicating the soil ecological function and measuring the effects of environmental governance methods ([48]). However, knowledge of soil bacterial response to exogenous N and P inputs is still limited, and the mechanisms that control the microbe-soil nutrient cycling remain elusive.
The recent development of high-throughput sequencing techniques ([31]) has overcome the intrinsic limitation of traditional microbial culturing techniques ([41]), thus improving the structural analysis of soil microbial communities ([44]) as well as identifying previously unknown microbial taxa ([5], [32]). With the development of techniques for structural analysis of soil microbial communities, high-throughput sequencing has revolutionized the field of soil microbial ecology. In particular, the 16S rRNA gene sequence, which contains highly conserved primer-binding sites and hypervariable, species-specific regions ([26]), is often used for the identification and classification of bacteria ([47]), providing a deep insight into the diversity of soil microbial communities. Additionally, the sequencing of bacterial 16S rRNA marker shows the potential for forest soil analysis. The spatial and temporal factors, which will undoubtedly come into play for soil comparisons, had little influence on soil traceability using this method ([23]). Although 16S rRNA sequencing was extensively used to characterize microbial communities of forest soils worldwide ([43], [16]), including China ([7]), it was rarely considered in relation to specific forest soil treatments, such as N and P fertilization ([59]).
In this study, we used high-throughput 16S rRNA sequencing method to explore the soil bacterial community diversity after N and P additions in a subtropical secondary Castanopsis sclerophylla forest. The objective was to test the effects of exogenous N and P inputs on forest soil properties and bacterial populations, and to determine the soil properties associated to the treatment-dependent compositional shifts of the local bacterial community. This investigation can strengthen our understanding of the effect of aggravated atmospheric deposition on soil microbial composition, and supplement the theoretical basis for the further study of the microbe-soil nutrient cycling mechanisms in subtropical forest ecosystem.
Materials and methods
Study sites
A field-simulated experiment was carried out from June 2011 to June 2015 in Jitan, Shitai County in the south part of Anhui Province, eastern China (30° 01′ 47″ N, 117° 21′ 23″ E - [30]). The overall climate in this region is a typical subtropical monsoon climate, with warm and humid summers, cold and dry winters. The study area is located at elevation from 100 to 150 m a.s.l. The mean annual temperature is 16 °C, with the highest of 38.8 °C and lowest of -8.9 °C. Mean annual precipitation is 1521 mm with uneven distribution, mainly from April to September ([30]). The soil in this area has been classified as yellow brown earth (Chinese Soil Taxonomic Classification) with acidic pH. Castanopsis sclerophylla was the main species with the dominance of 80- 90% at the study site. The stem density was 800-1189 trees hm-2 (DBH ≥ 5 cm), and the largest DBH was 30.6 cm. Other associated species were Liquidambar formosana and Pinus massoniana in the overstory and Loropetalum chinensis, Camellia cuspidata, Elaeagnus pungens and Lindera aggregata in the understory.
Experimental design
In 2011, a 43-years-old secondary Castanopsis sclerophylla forest was selected as experimental field. A moderate thinning has been conducted in this forest in 2008, and thereafter no management disturbances occurred. A randomized block design was used to avoid spatial heterogeneity. We chose three blocks with similar stand growths, species composition and site conditions to establish four treatments in each block: Control (CK), N addition (100 kg N ha-1 yr-1), P addition (50 kg N ha-1 yr-1), and N+P addition (100 kg N ha-1 yr-1 + 50 kg P ha-1 yr-1). In total, twelve 15 × 15 m plots were established with 8-10 m buffer strip between plots to avoid the interaction between treatments. N was added as NH4NO3 and P as Ca(H2PO4)2, which were sprayed near the soil surface at the end of each trimester from June 2011. All N and P treatments were mixed evenly with 20 L of stream water and sprayed by a hand-worked backpack sprayer, while each control plot received 20 L stream water with no N or P applications.
Soil sampling
Soil samples were collected on July 20, 2015. In preparation for sampling, we sterilized the sieve (2 mm) and the head of the handhold steel auger (6 cm internal diameter) in the high-pressure steam sterilization pot, and used the atmospheric-pressure steam sterilization pot for sterilizing the low-pressure polyethylene bags in laboratory the day before. Within each plot, five surface soil subsamples (0-10 cm) were randomly collected after removal of the litter layer, using the handhold steel auger. Totally 60 samples were collected into sterilized sealed polyethylene bags, and transported to the laboratory in a cooler box with ice bags. The samples were then passed through the sterilized sieve to remove roots and other debris, and divided into two equal parts. One was stored at 4 °C (fridge) until further analyses on soil physicochemical properties, and the other was combined into one composite sample for each plot, and then stored at -20 °C prior to genomic DNA extraction.
Soil physicochemical analysis
Soil water content (SWC) was measured by oven-drying at 105 °C to a constant weight. Soil pH and EC were determined using a pH/EC meter in 1:2.5 and 1:5 soil/water suspensions, respectively. Ammonium nitrogen (NH4+-N) and nitrate nitrogen (NO3--N) were measured by a flow injection auto-analyzer (FIAStar® 5000, FOSS, Denmark), as well as the total phosphorus (TP) after micro-Kjeldahl digestion. Soil organic carbon (SOC) and total nitrogen (TN) were measured with a CN Analyzer (EA 3000®, Vector, Italy). The C/N ratio was calculated as SOC divided by TN. Microbial biomass carbon (MBC) and nitrogen (MBN) were quantified using the chloroform fumigation-extraction method. Subsamples of fresh soil (30 g) were extracted with 100 mL of 0.5 M K2SO4 solution. Samples were shaken for 30 min and then filtered, while separate subsamples were fumigated with ethanol-free chloroform for 24 h at 25 °C and then extracted. Soil dissolved organic carbon (DOC - [24]) in extracts was immediately determined using a TOC auto-analyzer (Multi N/C 3100®, Jena Analytik, Germany). The difference in total N between extracts of fumigated and non-fumigated soils (NFE) was used for estimating MBN using the expression ([22]): MBN = NFE / 0.54 (mg kg-1). The difference in DOC content between fumigated and non-fumigated (CFE) extracts was used for MBC estimation using the expression ([56]): MBC = CFE / 0.45 (mg kg-1).
DNA extraction and 16S rRNA gene sequencing
We used the FastDNA™ SPIN Kit for soil (50 preps, MPbio, USA) to extract total DNA of soil microorganisms according to the manufacturer instructions. Afterward, using the 12 composite DNA samples as a template and the primer (319F 5’-ACTCCTACGGGAGGCAGCAG-3’; 806R 5’-GGACTACHVGGGTWTCTAAT-3’), we amplified the V3-V4 region of the bacterial 16S rRNA. All reactions were carried out in 25 µL (total volume) mixtures containing approximately 25 ng of genomic DNA extract, 12.5 µL PCR Premix, 2.5 µL of each primer, and PCR-grade water to adjust the volume. PCR reactions were performed in a Master cycler gradient thermocycler (Eppendorf, Hamburg, Germany) set to the following conditions: initial denaturation at 98 °C for 30 s; 35 cycles of denaturation at 98 °C for 10 s, annealing at 54/52 °C for 30 s, and extension at 72 °C for 45 s; and then final extension at 72 °C for 10 min. The PCR products were confirmed with 2% agarose gel electrophoresis. Throughout the DNA extraction process, ultrapure water, instead of a sample solution, was used to exclude the possibility of false-positive PCR results as a negative control. The PCR products were normalized by AxyPrepTM Mag PCR Normalizer (Axygen Biosciences, Union City, CA, USA), which allowed for the skipping of the quantifications step regardless of the PCR volume submitted for sequencing. The amplicon pools were prepared for sequencing with AMPure XT® beads (Beckman Coulter Genomics, Danvers, MA, USA) and with the Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, USA), respectively. PhiX Control library (Illumina) was combined with the amplicon library (expected at 30%). The total library was sequenced either on 300PE MiSeq runs using the standard Illumina sequencing primers ([9]).
Statistical analyses
According to the relative abundances, we divided the soil bacterial phyla into three parts: common phyla (>10%), rare common phyla (1-10%), and rare phyla (<1%). The operational taxonomic unit (OTU) was defined as a group of sequences with the same restriction profiles. Chao1 (SChao1) was determined according to methods described by Chao ([8]). The relative average abundance of phyla prevalent in soils from different treatments was calculated using the package Excel 2007® (Microsoft, Redwoods, CA, USA). Indices of bacteria biodiversity were estimated based on the OTUs using the methods of QIME ([6]), and Shannon-Wiener index (eqn. 1) and Simpson index (eqn. 2) were calculated as follows:
For bacterial diversity, n is the number of OTUs, and pi is the percentage of clones of the ith OTU, where S and N denote the total number of OTUs and number of clones in a library respectively, and pi = ni/N.
We used generalized linear mixed models (GLMMs) to evaluate the effect of N, P additions on the soil property and bacterial response variables. The bacterial response was set as the dependent variable, the main effect of block was built as the random effect, and the fixed effects consisted of both N and P treatments. The subplot replicates were regarded as a further random effect nested within the random effect of block to fit one of the soil properties as the dependent variable. Results were considered significant at P-value ≤ 0.05. The GLMMs analysis was conducted using the “lmer” function of the lme4 package ([4]) and the R statistical software (ver. 3.4.4 - ⇒ http://www.r-project.org/). Liner model was selected according to the Detrended Correspondence Analysis (DCA), and we used forward selection of variance inflation factors (VIF) to gradually filter the explanatory variables (rejecting the variables with VIF > 20). Consequently, 11 soil parameters were calculated in the redundancy analysis (RDA) for determining the relationship between environmental factors and the distribution of soil bacterial communities, represented by relative abundance of each OTUs. Monte Carlo permutation test was further applied to compute statistical significance (n = 999). The results of RDA analysis were visualized in the form of a biplot ordination diagram constructed by the R software.
Results
Soil physicochemical properties
As expected, soil physicochemical properties responded differently to N and P applications. N input had significant effect on the soil pH, EC, NH4+-N, NO3--N, DON, Ca, Mg (P < 0.001) and AP (P < 0.05), whereas P input affected several parameters significantly, such as SWC, pH, EC, NH4+-N, MBC, MBN, MBC/MBN, SOC, TP, AP, SOC/TN, SOC/TP, TN/TP and Ca (P < 0.05). Furthermore, the interaction effect of N:P had pronounced influence on the contents of soil pH, EC, NH4+-N, NO3--N, DON, MBN, TP and SOC/TN (Tab. 1). On the other hand, after N, P treatments, soils pH in the study area were more acidic, with pH dropping from 4.8 to 4.3 (see Tab. S1 in Supplementary material). The contents of pH, Mg, SOC/TP and TN/TP decreased significantly after N+P addition, whereas the contents of EC, NH4+-N, NO3--N, DOC, MBC, MBN, TP, K and AP increased obviously compared with CK (P < 0.05 - Tab. S1).
Tab. 1 - Outcome of the GLMMs model on the effect of N and P treatments and their interaction effect (N:P) on soil physicochemical properties.
| Soil property |
N | P | N:P | |||
|---|---|---|---|---|---|---|
| F | P | F | P | F | P | |
| SWC | 2.222 | 0.142 | 64.285 | <0.001 | 0.949 | 0.334 |
| pH | 80.227 | <0.001 | 29.407 | <0.001 | 11.175 | 0.002 |
| EC | 37.542 | <0.001 | 116.358 | <0.001 | 7.892 | 0.007 |
| NH4+-N | 48.218 | <0.001 | 48.988 | <0.001 | 14.792 | <0.001 |
| NO3--N | 64.986 | <0.001 | 0.251 | 0.618 | 41.510 | <0.001 |
| DOC | 2.761 | 0.102 | 3.610 | 0.063 | 3.072 | 0.085 |
| DON | 14.074 | <0.001 | 3.485 | 0.067 | 17.681 | <0.001 |
| MBC | 0.150 | 0.700 | 197.642 | <0.001 | 2.995 | 0.089 |
| MBN | 0.001 | 0.981 | 109.537 | <0.001 | 5.780 | 0.020 |
| MBC/MBN | 0.288 | 0.594 | 12.985 | 0.001 | 3.859 | 0.055 |
| SOC | 0.922 | 0.341 | 4.158 | 0.046 | 0.762 | 0.386 |
| TP | 0.793 | 0.378 | 95.887 | <0.001 | 14.264 | <0.001 |
| AP | 4.328 | 0.042 | 8.601 | 0.005 | 1.188 | 0.281 |
| TN | 0.963 | 0.331 | 1.912 | 0.172 | 0.725 | 0.398 |
| SOC/TN | 0.000 | 0.987 | 74.492 | <0.001 | 21.270 | <0.001 |
| SOC/TP | 0.015 | 0.903 | 33.686 | <0.001 | 0.101 | 0.752 |
| TN/TP | 0.001 | 0.982 | 11.728 | 0.001 | 2.327 | 0.135 |
| K | 1.881 | 0.178 | 0.011 | 0.916 | 2.299 | 0.137 |
| Ca | 29.217 | <0.001 | 89.478 | <0.001 | 8.217 | 0.006 |
| Mg | 29.696 | <0.001 | 0.333 | 0.566 | 2.104 | 0.153 |
Soil bacterial diversity
Based on the results of the GLMMs model, N and P treatments did not produce significant shifts in species richness, as well as in the tested diversity indices, like the number of observed species (OTUs), Shannon-Wiener index, Simpson index and Chao1 index (Tab. S2 in Supplementary material).
Bacterial community composition
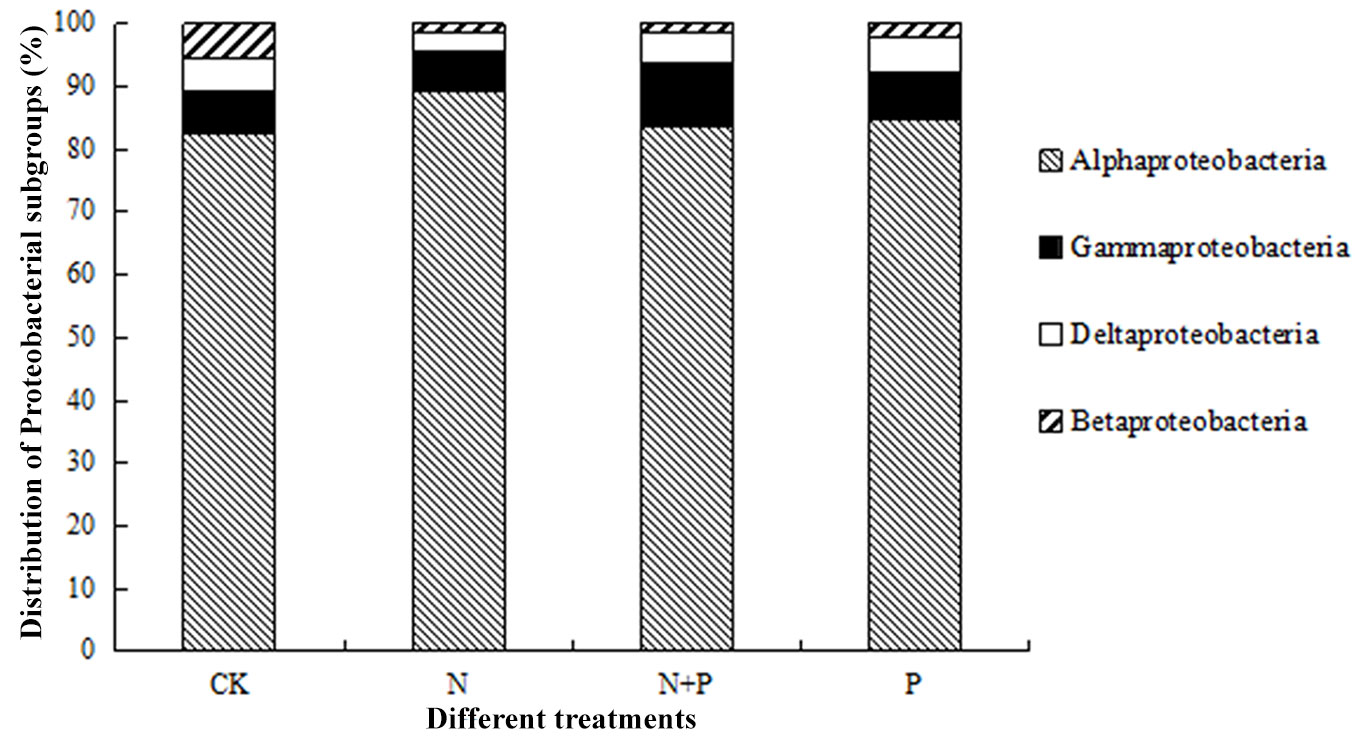
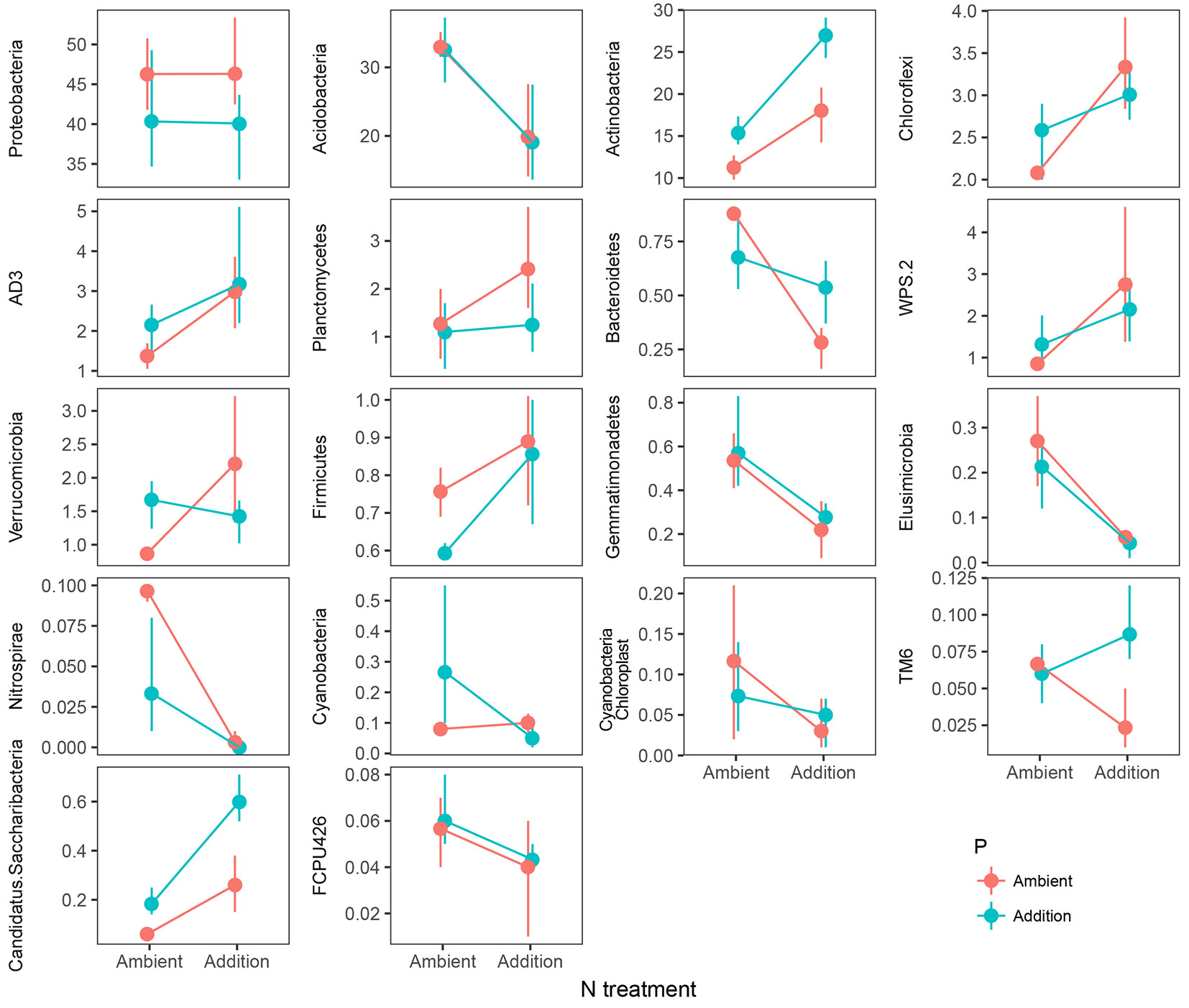
Proteobacteria-, Acidobacteria-, and Actinobacteria-associated OTUs accounted for more than 84% of the total, and these common phyla were the most abundant in both treatments and control (Tab. 2). The rare common phyla accounted for nearly 9% and the rare only took up less than 7% (Tab. 2). Proteobacteria was the most abundant bacterial phylum, and contained four primary classes out of six from the database of detected OTUs (α-, β-, γ- and δ-proteobacteria, number of OTUs > 5 - Fig. 1). Among the clone library of subgroup α-proteobacteria, the relative abundance decreased by 4.6% and 3.99% after N+P and P additions respectively, whereas increased by 3.15% after N additions. For γ-proteobacteria, the relative abundance decreased by 0.15% and 0.05% after N and P additions, whereas increased by 0.92% after N+P additions. Moreover, notable changes were observed among four treatments of ambient and addition for some other phyla (Fig. 2). As reported in Tab. 2 and Tab. S3 (Supplementary material), we found that N addition significantly (P < 0.05) reduced the relative abundances of specific taxonomic groups compared to the control, specifically for Acidobacteria (13.11%), Bacteroidetes (0.6%), Elusimicrobia (0.21%), Nitrospirae (0.1%) and TM6 (0.04%). The P addition significantly (P < 0.01) reduced the relative abundances of Nitrospirae (0.07%). Additionally, the GLMM analysis showed pronounced interactive effects of N and P additions (N:P) on phylum Nitrospirae (P < 0.01) and TM6 (P < 0.05 - Tab. S3 in Supplementary material).
Tab. 2 - Comparison of clone libraries of bacteria at phylum and class levels from forest soils at four different treatments. Number of OTUs > 5.
| Phylum | Class | Control (CK) | N | N+P | P | ||||
|---|---|---|---|---|---|---|---|---|---|
| OTUs | % | OTUs | % | OTUs | % | OTUs | % | ||
| Proteobacteria | (all) | 12606.50 | 46.28 | 7509.00 | 46.31 | 6237.67 | 40.07 | 8709.00 | 40.34 |
α-proteobacteria |
10349.00 | 38.15 | 6702.00 | 41.30 | 5218.00 | 33.55 | 7384.00 | 34.16 | |
γ-proteobacteria |
892.50 | 3.13 | 481.00 | 2.98 | 635.00 | 4.05 | 659.00 | 3.08 | |
δ-proteobacteria |
652.00 | 2.39 | 214.67 | 1.33 | 295.33 | 1.90 | 473.00 | 2.18 | |
β-proteobacteria |
710.00 | 2.60 | 111.00 | 0.69 | 88.00 | 0.56 | 192.67 | 0.91 | |
| Acidobacteria | - | 8459.50 | 32.97 | 3127.67 | 19.86 | 3145.33 | 19.09 | 6461.00 | 32.55 |
| Actinobacteria | - | 2818.50 | 11.25 | 2906.33 | 18.05 | 4229.67 | 26.99 | 3043.67 | 15.35 |
| Chloroflexi | - | 550.00 | 2.08 | 529.67 | 3.33 | 482.67 | 3.01 | 498.67 | 2.59 |
| AD3 | - | 328.50 | 1.37 | 479.00 | 2.99 | 492.00 | 3.18 | 400.33 | 2.15 |
| Planctomycetes | - | 263.50 | 1.27 | 379.00 | 2.41 | 211.33 | 1.25 | 180.33 | 1.10 |
| Bacteroidetes | - | 234.50 | 0.88 | 46.00 | 0.28 | 86.33 | 0.54 | 149.33 | 0.68 |
| WPS-2 | - | 234.00 | 0.85 | 436.33 | 2.76 | 353.67 | 2.16 | 258.67 | 1.32 |
| Verrucomicrobia | - | 223.00 | 0.87 | 345.67 | 2.21 | 229.67 | 1.42 | 317.33 | 1.67 |
| Firmicutes | - | 192.00 | 0.76 | 143.67 | 0.89 | 135.67 | 0.86 | 120.67 | 0.59 |
| Gemmatimonadetes | - | 153.50 | 0.54 | 36.67 | 0.22 | 42.67 | 0.28 | 131.33 | 0.57 |
| Elusimicrobia | - | 80.00 | 0.27 | 9.00 | 0.06 | 6.67 | 0.04 | 39.00 | 0.21 |
| Nitrospirae | - | 25.00 | 0.10 | 0.33 | 0.00 | 0.00 | 0.00 | 5.00 | 0.03 |
| Cyanobacteria | - | 22.50 | 0.08 | 15.67 | 0.10 | 8.67 | 0.05 | 41.00 | 0.27 |
| Chloroplast | - | 21.50 | 0.12 | 4.00 | 0.03 | 8.67 | 0.05 | 15.33 | 0.08 |
| TM6 | - | 17.00 | 0.07 | 4.33 | 0.03 | 13.67 | 0.09 | 11.67 | 0.06 |
| candidatus Saccharibacteria |
- | 15.00 | 0.06 | 41.33 | 0.26 | 95.00 | 0.60 | 41.33 | 0.18 |
| FCPU426 | - | 13.50 | 0.06 | 7.00 | 0.04 | 6.67 | 0.04 | 13.00 | 0.06 |
Fig. 1 - Proportions of four dominant Proteobacteria subgroups in the soil samples from different treatment plots.
Fig. 2 - Shifts of soil bacterial relative abundance at phylum level after N and P additions from different treatment plots. Error bars represent the standard error.
Various results were found among the bacteria when the genus level was considered. Compared with CK treatment (control), N addition significantly (P < 0.05) decreased by 81.56% the number of OTUs in the genus Burkholderia (Proteobacteria), by 72.39% in Gp2 (Acidobacteria), 100% in Ohtaekwangia (Bacteroidetes), 42.86% in Cohnella (Firmicutes) and 100% in Nitrospira in phylum Nitrospirae (Tab. S4 in Supplementary material). The N+P treatment significantly increased by 121.47% the number of OTUs in the genus Conexibacter (Actinobacteria), 533.33% in Saccharibacteria genera incertae sedis (candidatus Saccharibacteria), and mostly reduced the OTUs by 86.57% (P < 0.05) in the genus Burkholderia (Proteobacteria), 78.04% in Gp2 (Acidobacteria), 93.55% in Ohtaekwangia (Bacteroidetes) and 47.62% in Cohnella (Firmicutes). Moreover, P addition largely decreased (P < 0.05) the OTUs of Burkholderia in Proteobacteria (-69.34%), Gp3 in Acidobacteria (-52.39%), Cohnella in Firmicutes (-80.95%) and Nitrospira in Nitrospirae (-76.47%). Additionally, the genus Gp17 was not detected after all allochthonous additions, as well as some other genera not shown after specific treatment, such as the genera Hyphomicrobium, Variovorax, Gp10, Ohtaekwangia, Flavitalea and Nitrosospira.
Relationship between soil physicochemical properties and bacterial populations
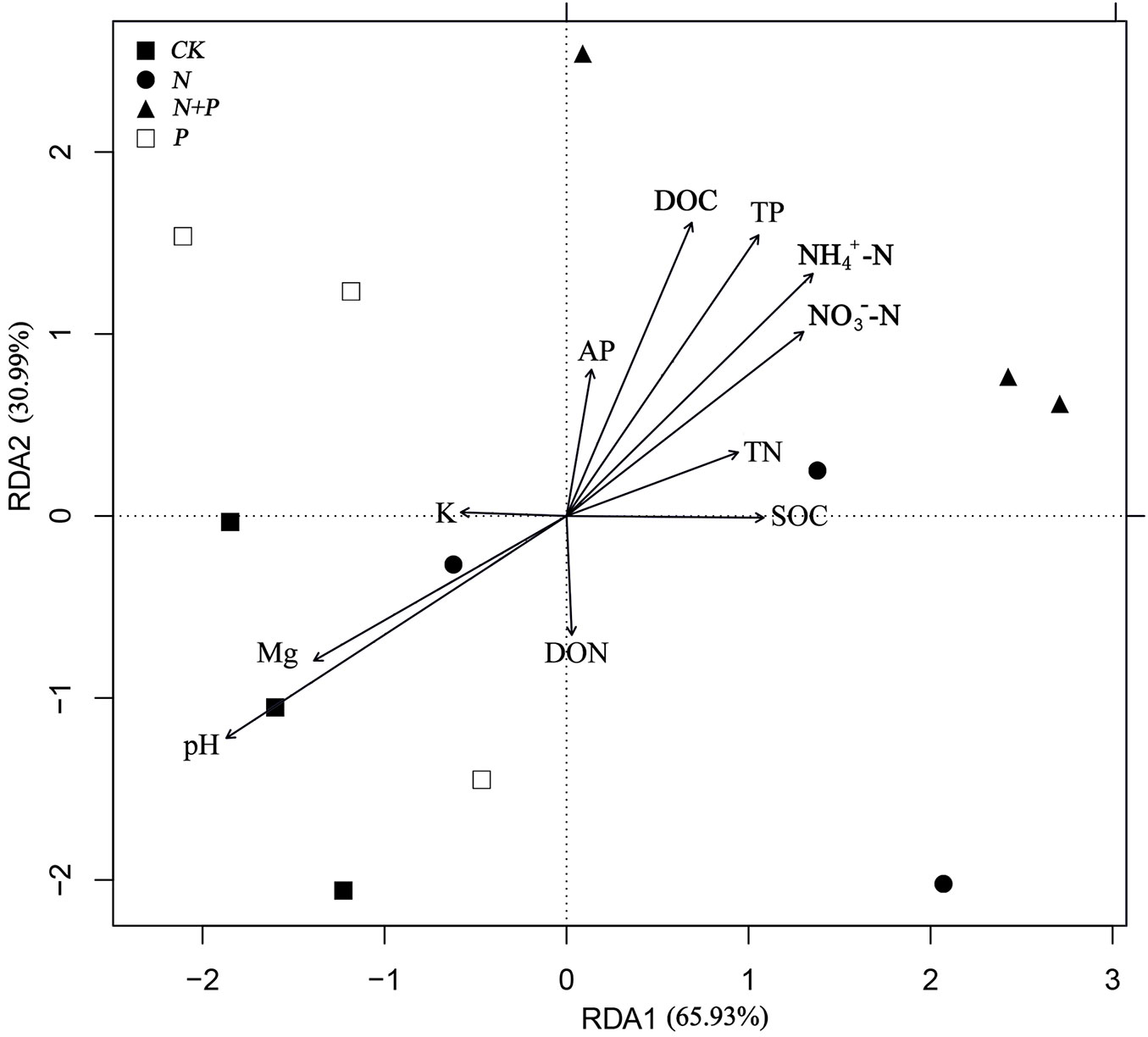
The RDA ordination plot shows the relationship between the phylum of bacterial populations and environmental variables (Fig. 3), which indicated that the plots subjected to the four treatments were clearly separated from each other. Altogether, the soil physicochemical properties explained 96.92% of the total variation by first two axes, and pH best explained the differences between treatments, which was validated by Monte Carlo permutation tests (P = 0.015 - Tab. S5 in Supplementary material).
Fig. 3 - Ordination plot of the results from the redundancy analysis (RDA). The plot was used to explore the relationships between selected soil properties and bacterial community composition. Numbers in brackets represent the percentage of variation of the data explained by each factor, RDA1 explains 65.93% of the total variation and RDA2 explains 30.99% of the total variation, respectively. The primary factors for the variables of the environmental data are represented by arrows.
Discussion
Effects of N and P additions on soil parameters
Soil pH in N, P and N+P treatments were significantly more acidic than in control plots (Tab. S1 in Supplementary material), suggesting that N, P additions accelerated soil acidity. Similar to our result, Tian et al. ([52]) reported a decreased pH value after 4-year nitrogen fertilization in Guniujiang (GNJ, Anhui province), southern China, especially at 0-10 cm soil depth, in subtropical evergreen broad-leaf forests where the value of soil pH was below 5. The recent paper by Zheng et al. ([59]) about the specific effect of long-term N and P fertilization of east China forest soils, addressed the topic of the effect on inorganic P solubilizing bacteria (iPSB) in alkaline soils. Its result showed that long-term N addition inhibited iPSB by reducing soil pH, eventually increasing soil P release. Although our sampling sites belongs to acid soil, the pH after N and P additions decreased and the contents of TP and AP increased significantly, which suggests that IPSB populations may modulate the acidification and P release in the soil. In general, after N saturation, excessive N inputs may lead to N supply exceeding the demand of soil microorganism and plant, causing the NO3--N loss through leaching and denitrification, which may accelerate the process of soil acidification ([49]). Tian et al. ([51]) found that N addition suppressed microbial biomass in subtropical forest where N is close to saturation, and excess N to these forests may lead to a high risk of N saturation and aggravated P limitation, which might further negatively affect the survival of microbial communities. On the other hand, Zhang et al. ([57]) observed P alleviating the nutrient limitation and soil acidification in the Chinese fir plantations in subtropical China, which was not reflected by our findings. This may be due to the different site conditions, including tree species (evergreen broadleaved vs. coniferous forest), soil types (yellow brown vs. red soil) and other factors.
Effects of N and P additions on bacterial populations
As reviewed by Janssen ([21]), the phylum Proteobacteria comprises the largest fraction of the bacterial community in soil ecosystems. Generally, the subgroup α-Proteobacteria grows at relative lower levels of nutrients ([11]). In our study, GLMMs analysis showed no significant difference within the class of α-Proteobacteria after N and P additions. At the genus level, however, a remarkable difference came out after nutrient additions. For example, the OTUs numbers of genus Rhizobium (α-Proteobacteria) decreased significantly after N and N+P addition (P < 0.05), and Nitrospirillum and Nitrobacter (α-Proteobacteria) decreased after N+P addition (Tab. S4). These indicate that our site may be N enriched and the large proportion of α-Proteobacteria may be limited by copiotrophic attributes, consistent with previous studies that α-Proteobacteria possibly had an oligotrophic affinity and was adapted to oligotrophy and acidic soils ([12], [38]). On the other hand, significant decreases were detected between treatments and control in the subgroup β-Proteobacteria. Its genus Burkholderia, which showed the largest abundance in all samples and had significant differences between treatments, is known to include efficient mineral-weathering and nitrogen-fixing bacteria ([28]). In our study, however, its relative abundance decreased significantly (P < 0.05) after additions compared with control (Tab. S4 in Supplementary material). This may be due to the relatively high level of nutrient enrichment that increases soil nitrogen availability and weakens bacterial competitive advantage. Previous study has proved that biological nitrogen fixation is an energy-intensive process and requires a suitable reductant to support electron transport to nitrogenase ([45]). In the case of high levels of available N in the soil, the gene for nitrogen fixation will usually not express ([45]), which may negatively affect the growth of N-fixing bacteria. Additionally, in assessing the effect of N addition on nutrient cycling processes, bacterial and archaeal nitrification and denitrification gain importance after N saturation, and initiated the nutrient cycling, resulting in the efflux of N compounds from the soil ecosystem ([1]). Therefore, bacteria with more efficient nitrification pathways were more likely to be selected ([13]), which may lead to higher rate of nitrification despite the reduced abundance of nitrifiers and genes related to N cycling processes.
Acidobacteria, which is known to grow under oligotrophic and acidic environment, is physiologically diverse and ubiquitous, especially in soil. However, it has been reported that some members of Acidobacteria did not exist in the soil at relative lower pH ([3]). Jones et al. ([25]) observed that the effect of pH on multiple members of Acidobacteria was not the same. This likely explain why the abundance of Acidobacteria did not increase with the decrease of soil pH in our study. Further analysis indicated that the response of different subgroups of Acidobacteria to soil pH was varied: the relative abundances of genera Gp1, 2, 3, 12, 13 and 15 were negatively correlated with soil pH, while Gp4, 6, 7, 10, 11, 16, 17, 18, 22 and 25 showed positive correlations ([25]). Specifically, we observed that the abundance of Gp2, 3 and 6 were the predominant genera with significant differences to treatments, and showed the similar decreasing trend with soil acidification (Tab. S4). This result is partly in accordance with the findings by Zhang et al. ([58]), who reported that the relative abundances of genera Gp1, 2 and 3 were significantly and positively correlated with soil pH, while others presented opposite patterns in the Shennongjia Mountain. Some studies also revealed that other soil parameters, such as mineral element contents ([39]), soil enzyme activity ([58]), and plant diversity ([38]) were closely related to the abundances of certain genera, while the contribution of these factors on the change of the community composition needs to be further investigated.
Conclusions
Overall, exogenous N and P inputs did not produce significant changes to the bacterial species richness, as well as to the diversity indices, which indicate a relative balanced soil microbial community structure able to cope with external environmental shifts. On the other hand, soil bacterial populations after N and P treatments clearly differed compared with control sites, especially for several subgroups of the phyla Proteobacteria and Acidobacteria. Inorganic nutrient additions alter the status of nutrient enrichment or limitation, which may lead to variations of soil physico-chemical properties, as well as microbial quantity and activity. Moreover, soil pH, as an overriding factor on determining bacterial community structure, closely affected the distribution of bacterial communities, which provides an insight to investigate soil microbial shifts mediated largely by microbe-soil interactions in the subtropical forest ecosystem.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, No. 31770672 and 31370626) and the National Basic Research Program of China (973 Program, No. 2010CB950602). We gratefully acknowledge Mr. ZF Wang and J Xu for the field sampling, and Dr. C Shi and Mr. T Xie for assistance with molecular biology experiments.
References
Gscholar
Gscholar
Gscholar
Gscholar
CrossRef | Gscholar
Online | Gscholar
Authors’ Info
Authors’ Affiliation
Jingjing Wang
Xinlei Fu
Xiaoniu Xu
School of Forestry and Landscape Architecture, Anhui Agricultural University, Hefei 230036 (China)
College of Life and Environment Sciences, Huangshan University, Huangshan 245041 (China)
Corresponding author
Paper Info
Citation
Teng Z, Cui J, Wang J, Fu X, Xu X (2018). Effect of exogenous nitrogen and phosphorus inputs on the microbe-soil interaction in the secondary Castanopsis sclerophylla forest in east China. iForest 11: 794-801. - doi: 10.3832/ifor2673-011
Academic Editor
Giorgio Alberti
Paper history
Received: Nov 02, 2017
Accepted: Oct 16, 2018
First online: Dec 14, 2018
Publication Date: Dec 31, 2018
Publication Time: 1.97 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2018
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 45895
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 37713
Abstract Page Views: 3916
PDF Downloads: 3284
Citation/Reference Downloads: 14
XML Downloads: 968
Web Metrics
Days since publication: 2611
Overall contacts: 45895
Avg. contacts per week: 123.04
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2018): 8
Average cites per year: 1.00
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Response of soil bacterial communities to nitrogen and phosphorus additions in an age-sequence of subtropical forests
vol. 14, pp. 71-79 (online: 11 February 2021)
Research Articles
Seasonal dynamics of soil respiration and nitrification in three subtropical plantations in southern China
vol. 9, pp. 813-821 (online: 29 May 2016)
Research Articles
The manipulation of aboveground litter input affects soil CO2 efflux in a subtropical liquidambar forest in China
vol. 12, pp. 181-186 (online: 10 April 2019)
Research Articles
Soil respiration along an altitudinal gradient in a subalpine secondary forest in China
vol. 8, pp. 526-532 (online: 01 December 2014)
Research Articles
Effects of tree species, stand age and land-use change on soil carbon and nitrogen stock rates in northwestern Turkey
vol. 9, pp. 165-170 (online: 18 June 2015)
Research Articles
The effect of clear-cut age on soil organic carbon and nitrogen indices in Scots pine (Pinus sylvestris L.) stands
vol. 18, pp. 146-153 (online: 09 June 2025)
Short Communications
Not all long-distance-exploration types of ectomycorrhizae are the same: differential accumulation of nitrogen and carbon in Scleroderma and Xerocomus in response to variations in soil fertility
vol. 14, pp. 48-52 (online: 18 January 2021)
Research Articles
Soil stoichiometry modulates effects of shrub encroachment on soil carbon concentration and stock in a subalpine grassland
vol. 13, pp. 65-72 (online: 07 February 2020)
Research Articles
Fungal and bacterial communities in a forest relict of Pinus pseudostrobus var. coatepecensis
vol. 16, pp. 299-306 (online: 09 November 2023)
Short Communications
Seasonal change in soil nitrogen mineralization in young Chamaecyparis obtusa stands at the upper and lower positions on a slope in central Japan
vol. 18, pp. 197-201 (online: 20 July 2025)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword