Differentiation of Populus species by chloroplast SNP markers for barcoding and breeding approaches
iForest - Biogeosciences and Forestry, Volume 8, Issue 4, Pages 544-546 (2014)
doi: https://doi.org/10.3832/ifor1326-007
Published: Nov 13, 2014 - Copyright © 2014 SISEF
Short Communications
Collection/Special Issue: COST Action FP0905
Biosafety of forest transgenic trees and EU policy directives
Guest Editors: Cristina Vettori, Matthias Fladung
Abstract
About 30 species within the genus Populus are classified in six sections. Several species belonging to different sections are cross-compatible, resulting in a high number of naturally occurring interspecies hybrids. Additionally, an even higher number of hybrids has been produced in huge breeding programs during the last 100 years. Hence, determination of poplar species used for production of “multi-species-hybrids” is often difficult and, therefore, a challenge in developing molecular markers for species identification. Fourteen out of the 30 poplar species known are used more or less regularly for production of artificial hybrids and clones. In this study, we focused on over 20 chloroplast regions, and we tested 23 primer combinations already established for “Barcoding” approaches and seventeen new primer combinations designed earlier for the applicability to differentiate fourteen poplar species. In contrast to the self-designed primer combinations with a much higher amplification success, only about half of the established barcoding primer combinations yielded amplification products. In total, for eleven of the fourteen used poplar species we found species-specific SNPs or Indels. Most of the variation was found in intergenic spacers. In order to design an inexpensive and fast method of species identification, we developed PCR-RFLPs applicable for seven of the species-specific SNPs. Altogether there is high variation in chloroplast intergenic spacers within the genus Populus, illustrated by the fact that four primer combinations are needed to differentiate eleven species. Thus, we support the suggestion of using multi-locus combinations in barcoding analyses.
Keywords
Chloroplast Genome, SNPs, Indel, Barcoding, Intergenic Spacer
Introduction
The genus Populus is one of the world’s most important tree genera. High growth rates, particularly of some interspecies hybrids, and a broad applicability ranging from wood and paper to energy production, led to their widespread cultivation in Europe and North America ([2]). Hybrids between various Populus species belonging to the same or different sections are commonly used in short rotation coppices (SRC) for biomass production. Further, clones of various poplar species are easily accessible for genetic transformation, and a huge number of transgenic poplar clones have been tested in the field for biosafety reasons and commercial application ([4], [14]).
Especially interspecies-hybrids are well suited for biomass production because of their superior growth and advanced resistance traits. However, due to extreme variability of Populus hybrids, the species identification within the genus Populus using morphological characters sometimes proved to be difficult. A high number of interspecies-hybrids has been artificially produced in extensive breeding programs during the last 100 years ([12]), in addition to naturally occurring hybrids also possibly used for breeding purposes. The many years of breeding work, including repetitive crosses and back-crosses, have caused a lack of background information for many of commercially available hybrid clones originally used, though clear species identification is inevitably necessary for breeding activities aimed at licensing new high-efficiency clones.
Due to its small genome size, poplar has been selected as a model species for tree genomics, resulting in the publication of the full genome of western black cottonwood, P. trichocarpa ([13]). Using this genomic information, we evaluated and tested the usability of both already published plant barcoding regions from the chloroplast ([5], [1], Barcode of Life: ⇒ http://www.barcodeoflife.org/) and novel developed primers by Schroeder et al. ([11]) for their efficacy in differentiating the 14 poplar species listed in Tab. 1.
Tab. 1 - Numbers of species-specific SNPs, number of restriction sites within SNPs, and Indels for 14 Populus species in 24 chloroplast fragments. Labels for each species are given in brackets.
| Section | Species | No. of individuals |
No. SNPs |
No. restriction sites |
No. Indels |
|---|---|---|---|---|---|
| Populus | P. alba (alb) | 26 | 4 | 1 | 3 |
| P. tremula (tre) | 40 | 14 | 3 | 7 | |
| P. tremuloides (tro) | 40 | 31 | 4 | 11 | |
| Aigeiros | P. nigra (nig) | 36 | 7 | 2 | 3 |
| P. deltoides (del) | 19 | 9 | 1 | 4 | |
| Tacamahaca | P. trichocarpa (tri) | 35 | 6 | 1 | 1 |
| P. maximowiczii (max) | 18 | 2 | 0 | 1 | |
| P. simonii (sim) | 9 | 2 | 0 | 1 | |
| P. koreana (kor) | 4 | 0 | 0 | 0 | |
| P. cathayana (cat) | 1 | 0 | 0 | 0 | |
| P. szechuanica (sze) | 1 | 2 | 1 | 0 | |
| P. ussuriensis (uss) | 1 | 0 | 0 | 0 | |
| Leucoides | P. wilsonii (wil) | 1 | 4 | 0 | 3 |
| Turanga | P. euphratica (eup) | 1 | 11 | 0 | 2 |
Material and Methods
Sample information
Species and number of individuals (genotypes) per species used for this study are listed in Tab. 1. Individuals of three Populus species of the section Populus (P. alba, P. tremula, P. tremuloides) were collected in the arboretum and were provided by colleagues from Bavaria and Saxony, while P. euphratica (section Turanga) has been taken from the tissue culture of the Thünen-Institute of Forest Genetics in Grosshansdorf (Germany). For the two European species, further individuals have been sampled by colleagues all over the distribution area. Some of the individuals of Populus spp. of the sections Aigeiros (P. nigra and P. deltoides) and Tacamahaca (P. trichocarpa, P. maximowiczii, P. cathayana, P. szechuanica, P. ussuriensis, P. koreana) were kindly provided by the NW-FVA (Nordwestdeutsche Forstliche Versuchsanstalt) in Hann. Muenden (Germany). Six individuals of P. nigra were from the Ural Mountains and others were taken as samples at riversides in Germany. Nine individuals of P. maximowiczii and seven individuals of P. simonii have been provided by the Thünen-Institute of Forest Genetics in Waldsieversdorf (Germany). Several individuals of some of the above mentioned species (among others P. wilsonii) were received from the Botanical Gardens of Hamburg, Marburg, Tuebingen and Dresden (Germany).
DNA extraction and PCR amplification
One cm2 of a single leaf was ground to powder in liquid nitrogen. Total DNA was extracted, following a modified ATMAB protocol by Dumolin et al. ([3]). A standard protocol was used for PCR reactions ([11]). The PCR reactions contained from 20 to 50 ng template DNA, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 1.8 mM MgCl2, 200 μM dNTPs, 1 unit Taq polymerase, and 0.4 μM of each primer in a total volume of 25 μl. PCR was carried out in a Biometra Personal Thermocycler (Goettingen, Germany) with a pre-denaturation step at 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, suitable annealing temperature for each primer combination (ranging between 50°C and 65°C) for 45 sec or 1 min, 72°C for 1 min and a final elongation at 72°C for 10 min. PCR amplification products were checked on a 1 % agarose gel stained with Roti-Safe GelStain (Carl Roth GmbH & Co. KG, Karlsruhe, Germany).
Tested chloroplast regions
Twenty three published barcoding primer combinations were used for PCR amplification of coding and non-coding (intergenic spacers) chloroplast regions. Additionally, 17 primer combinations were newly designed taking advantage of the sequence of the Populus trichocarpa chloroplast genome ([9], [11]). The primer combinations are given in detail in Schroeder et al. ([11]).
Sequencing and PCR-RFLPs
For sequencing, 15 µl of the PCR product was purified either with 5 µl of 4 M lithium chloride or using the “High Pure PCR Product Purification” kit from Roche (Mannheim, Germany). All the sequences were provided by StarSeq (Mainz, Germany).
Up to ten individuals per species and primer combination have been sequenced. Obtained sequences were aligned and screened for the presence of SNPs by using either the software SFRMAO 7.1.0 from DNAStar (Madison, USA) or SFRVFODIFS 4.9 (Gene Codes Corporation, Ann Arbor, USA). Differentiating sequences (SNPs, Indels) were checked for a possible application of restriction enzymes using the software NEBDVUUFS V2.0 from New England BioLabs Inc (Ipswich, USA). Individuals already sequenced, plus further individuals given in Tab. 1, have been used to test the usability of PCR-RFLPs for species differentiation.
Results and iscussion
Twelve of the 23 used barcoding primer combinations and 16 out of the 17 newly-designed primer combinations yielded PCR amplification products in all the individuals of the 14 species tested. Twenty-four of these amplification products have been sequenced and checked for species-specific SNPs or Indels for the first seven species in Tab. 1 (P. alba, P. tremula, P. tremuloides, P. nigra, P. deltoides, P. trichocarpa, P. maximowiczii), and 19 fragments have been sequenced for the remaining seven species (P. simonii, P. koreana, P. cathayana, P. szechuanica, P. ussuriensis, P. wilsonii, P. euphratica). Species-specific SNPs or Indels were not found only in three species (P. koreana, P. cathayana and P. ussuriensis - Tab. 1). For these species, five further primer combinations were tested. In addition, more individuals of the last five species listed in Tab. 1 (P. cathayana, P. szechuanica, P. ussuriensis, P. wilsonii and P. euphratica) are needed to validate the species-specificity of the genetic variation.
Interestingly, for the two species P. wilsonii and P. euphratica, each belonging to two different sections, comparatively many SNPs and Indels have been found than for most of the other species of the sections Tacamahaca and Aigeiros (Tab. 1). Also, the similarities between species within the sections are different. No genetic variation could be found between the two species P. koreana and P. cathayana. These two species showed a similarity to P. maximowiczii (so far differing only by one SNP) much higher than to all the other species within the section Tacamahaca. Analysis of kinship with a wide range of markers for all the species could be of interest.
No species-specific SNPs or Indels were found by the analysis of three chloroplast regions (atpF-atpH, rps2-rpoC2 and psbJ-psbF) in the first seven species listed in Tab. 1, though the same chloroplast regions revealed the presence of SNPs or Indels in further seven species. The highest number of species-specific SNPs and Indels were found within the intergenic spacers trnV-atpE, matK-trnK, psaJ-rpl33, trnH-psbA, psbK-psbI and atpB-rbcL, followed by the coding region rpoC (Tab. 2). Four primer combinations were needed for the differentiation of 11 species with species-specific SNPs or Indels. Based on our results, we therefore recommend the use of a multi-locus combination of the plastid genome to increase barcoding potential, according to several previous reports ([5], [6], [7], [8]).
Tab. 2 - Number of polymorphisms, SNPs, and Indels detected in PCR amplicons for all successful amplified primer combinations. (Length): length of the PCR product amplified with the given primers in base pairs referred to P. trichocarpa; (No. Poly): total number of polymorphisms (SNPs and Indels): (No. SNPs): species-specific SNPs identified; (No. Indels): species-specific Indels identified; (% var): percent of variable sites calculated as the ratio between the total number of polymorphisms and the sequence length.
| Chloroplast region |
Length (bp) |
No. Poly |
No. SNPs |
No. Indels |
% var |
|---|---|---|---|---|---|
| trnV-atpE | 740 | 32 | 18 | 8 | 4.3 |
| psaJ-rpl33 | 617 | 12 | 6 | 3 | 1.9 |
| rbcL-accD | 762 | 12 | 4 | 1 | 1.6 |
| matK-trnK | 851 | 11 | 7 | 0 | 1.3 |
| trnC-petN | 441 | 11 | 5 | 1 | 2.5 |
| rpoC2-rpoC1 | 533 | 10 | 5 | 1 | 1.9 |
| trnG-psbK | 545 | 10 | 3 | 1 | 1.8 |
| psbK-psbI | 593 | 8 | 6 | 0 | 1.3 |
| trnH-psbA | 367 | 8 | 4 | 2 | 2.2 |
| rps8-rpl14 | 455 | 7 | 2 | 2 | 1.5 |
| ndhE-ndhG | 356 | 7 | 2 | 2 | 2.0 |
| rpoB2-rpoB4 | 548 | 7 | 4 | 0 | 1.3 |
| rbcLa-rbcLajf634R | 654 | 7 | 2 | 0 | 1.1 |
| 3F_KIM-1R_KIM (matK) | 882 | 6 | 4 | 1 | 0.7 |
| rps4_trnL | 829 | 5 | 3 | 0 | 0.6 |
| atpI-rps2 | 570 | 4 | 3 | 0 | 0.7 |
| rps2-rpoC2 | 524 | 4 | 0 | 0 | 0.8 |
| ycf10-petA | 359 | 3 | 0 | 1 | 0.8 |
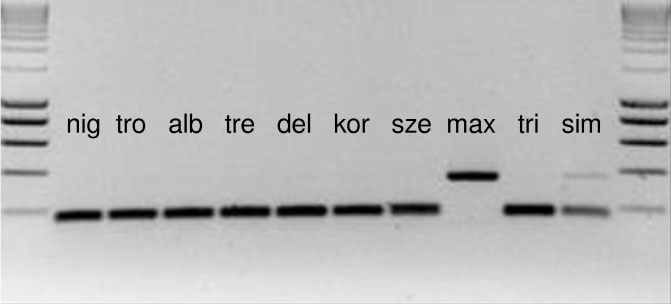
SNPs and Indels identified in this study could be an important contribution to the project “barcoding of life”. Their efficient application in the identification of Populus species for breeding purposes requires rapid test methods to be developed, in order to avoid sequencing or the use of capillary electrophoresis. To this purpose, all the sequences with SNPs were checked for restriction sites for use as PCR-RFLPs. Suitable restriction sites were found for 13 SNPs in seven species (Tab. 1). Some further restriction sites offered the possibility of a negative identification, that means all species can be excluded but one. An example of a PCR-RFLP is given in Fig. 1, where P. maximowiczii is characterized by the lack of a specific restriction site.
Fig. 1 - PCR-RFLP of the intergenic spacer ndhE-ndhG. P. maximowiczii (max) shows a larger PCR product due to the lack of the restriction site, whereas all other species were cut into two fragments of nearly the same size. Size marker (first and last lanes) is the SMART ladder (Eurogentec, Belgium). For species’ labels, see Tab. 1.
Conclusions
In this study, we demonstrated the applicability of SNP markers in breeding programs. The combination of SNPs with PCR-RFLPs and length polymorphism provides an easy-to-use, fast, and low cost method for identifying and distinguishing closely-related poplar species.
We recommend the use of intergenic spacers for differentiating closely related species within the genus Populus because of the higher overall variability ([10], [11]).
Acknowledgments
This project is supported by the Federal Ministry of Food and Agriculture via the “Fachagentur Nachwachsende Rohstoffe e.V.” (FNR). We would like to thank the greenhouse staff for harvesting of leaves from high poplar trees, our technical assistant Susanne Bein for the laboratory work, and the colleagues providing us with plant material. Our special thanks go to Dietrich Ewald and Georg von Wühlisch for helping us with their contacts to colleagues from China and the USA, and to Dina Führmann for proof reading of the English grammar and style. We are grateful to the Botanical Gardens from Hamburg, Marburg, Dresden and Tübingen for supplying us with poplar reference material.
References
Gscholar
CrossRef | Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Matthias Fladung
Thünen Institute of Forest Genetics, Sieker Landstrasse 2, D-22927 Grosshansdorf (Germany)
Corresponding author
Paper Info
Citation
Schroeder H, Fladung M (2014). Differentiation of Populus species by chloroplast SNP markers for barcoding and breeding approaches. iForest 8: 544-546. - doi: 10.3832/ifor1326-007
Academic Editor
Elena Paoletti
Paper history
Received: Apr 28, 2014
Accepted: Sep 16, 2014
First online: Nov 13, 2014
Publication Date: Aug 02, 2015
Publication Time: 1.93 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2014
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 55436
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 46543
Abstract Page Views: 3386
PDF Downloads: 4055
Citation/Reference Downloads: 45
XML Downloads: 1407
Web Metrics
Days since publication: 4136
Overall contacts: 55436
Avg. contacts per week: 93.82
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2015): 11
Average cites per year: 1.00
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Chloroplast DNA barcoding genes matK and psbA-trnH are not suitable for species identification and phylogenetic analyses in closely related pines
vol. 15, pp. 141-147 (online: 25 April 2022)
Research Articles
Identification and molecular characterization of LTR and LINE retrotransposable elements in Fagus sylvatica L.
vol. 2, pp. 119-126 (online: 10 June 2009)
Research Articles
Chloroplast microsatellites as a tool for phylogeographic studies: the case of white oaks in Poland
vol. 8, pp. 765-771 (online: 19 July 2015)
Research Articles
Comparison of range-wide chloroplast microsatellite and needle trait variation patterns in Pinus mugo Turra (dwarf mountain pine)
vol. 10, pp. 250-258 (online: 11 February 2017)
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Genetic diversity of core vs. peripheral Norway spruce native populations at a local scale in Slovenia
vol. 11, pp. 104-110 (online: 31 January 2018)
Review Papers
Genetic diversity and forest reproductive material - from seed source selection to planting
vol. 9, pp. 801-812 (online: 13 June 2016)
Research Articles
Age trends in genetic parameters for growth and quality traits in Abies alba
vol. 9, pp. 954-959 (online: 07 July 2016)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword