Differentiation of Populus species by chloroplast SNP markers for barcoding and breeding approaches
Hilke Schroeder , Matthias Fladung
iForest - Biogeosciences and Forestry, Volume 8, Issue 4, Pages 544-546 (2014)
doi: https://doi.org/10.3832/ifor1326-007
Published: Nov 13, 2014 - Copyright © 2014 SISEF
Short Communications
Collection/Special Issue: COST Action FP0905
Biosafety of forest transgenic trees and EU policy directives
Guest Editors: Cristina Vettori, Matthias Fladung
Abstract
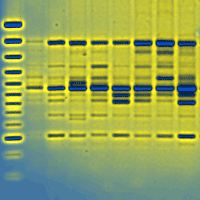
About 30 species within the genus Populus are classified in six sections. Several species belonging to different sections are cross-compatible, resulting in a high number of naturally occurring interspecies hybrids. Additionally, an even higher number of hybrids has been produced in huge breeding programs during the last 100 years. Hence, determination of poplar species used for production of “multi-species-hybrids” is often difficult and, therefore, a challenge in developing molecular markers for species identification. Fourteen out of the 30 poplar species known are used more or less regularly for production of artificial hybrids and clones. In this study, we focused on over 20 chloroplast regions, and we tested 23 primer combinations already established for “Barcoding” approaches and seventeen new primer combinations designed earlier for the applicability to differentiate fourteen poplar species. In contrast to the self-designed primer combinations with a much higher amplification success, only about half of the established barcoding primer combinations yielded amplification products. In total, for eleven of the fourteen used poplar species we found species-specific SNPs or Indels. Most of the variation was found in intergenic spacers. In order to design an inexpensive and fast method of species identification, we developed PCR-RFLPs applicable for seven of the species-specific SNPs. Altogether there is high variation in chloroplast intergenic spacers within the genus Populus, illustrated by the fact that four primer combinations are needed to differentiate eleven species. Thus, we support the suggestion of using multi-locus combinations in barcoding analyses.
Keywords
Chloroplast Genome, SNPs, Indel, Barcoding, Intergenic Spacer
Authors’ Info
Authors’ address
Matthias Fladung
Thünen Institute of Forest Genetics, Sieker Landstrasse 2, D-22927 Grosshansdorf (Germany)
Corresponding author
Paper Info
Citation
Schroeder H, Fladung M (2014). Differentiation of Populus species by chloroplast SNP markers for barcoding and breeding approaches. iForest 8: 544-546. - doi: 10.3832/ifor1326-007
Academic Editor
Elena Paoletti
Paper history
Received: Apr 28, 2014
Accepted: Sep 16, 2014
First online: Nov 13, 2014
Publication Date: Aug 02, 2015
Publication Time: 1.93 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2014
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 55229
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 46456
Abstract Page Views: 3305
PDF Downloads: 4027
Citation/Reference Downloads: 45
XML Downloads: 1396
Web Metrics
Days since publication: 4108
Overall contacts: 55229
Avg. contacts per week: 94.11
Citation Metrics
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2015): 11
Average cites per year: 1.00
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
References
The culture of poplars in Eastern North America. Department of Forestry, Michigan State University, East Lansing, MC, USA, pp. 168.
Gscholar
Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources 9: 439-457.
CrossRef | Gscholar
Biology of Populus and its implications for management and conservation. NRC Research Press. Montreal, Canada, pp. 542.
Gscholar