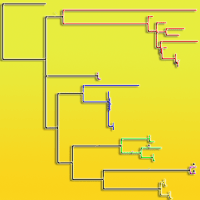
The largest and most economically important conifer genus Pinus is widespread in the northern hemisphere. Comprehensive phylogenies relying on complete chloroplast gene sequences are now available for the entire genus. However, phylogenetic relationships remain unresolved for certain lineages. One such example, which is also inconsistent in terms of biogeography, is within the subsection Pinus and includes five taxa: Pinus densiflora, P. nigra, P. resinosa, P. sylvestris and P. mugo / uncinata species complex. In this study, we use this clade as an example to explain weak support in phylogenetic studies of closely related pine species and show that some of the most popular genetic markers, namely the chloroplast DNA barcoding sequences matK, psbA- trnH and rbcL, are not recommended for species identification purposes in European pines. In addition, we show that matK and psbA-trnH contain contradicting phylogenetic signals in some of the most economically important pine species.
Keywords
, , ,
Citation
Olsson S, Giovannelli G, Roig A, Spanu I, Vendramin GG, Fady B (2022). Chloroplast DNA barcoding genes matK and psbA-trnH are not suitable for species identification and phylogenetic analyses in closely related pines. iForest 15: 141-147. - doi: 10.3832/ifor3913-015
Academic Editor
Alberto Santini
Paper history
Received: Jun 25, 2021
Accepted: Feb 15, 2022
First online: Apr 25, 2022
Publication Date: Apr 30, 2022
Publication Time: 2.30 months
© SISEF - The Italian Society of Silviculture and Forest Ecology 2022
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.

Breakdown by View Type
(Waiting for server response...)
Article Usage
Total Article Views: 35655
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 30052
Abstract Page Views: 3063
PDF Downloads: 2021
Citation/Reference Downloads: 9
XML Downloads: 510
Web Metrics
Days since publication: 1410
Overall contacts: 35655
Avg. contacts per week: 177.01
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2022): 2
Average cites per year: 0.50
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
(1)
Alvaréz I, Wendel JF (2003)Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogeny and Evolution 29: 417-434.
CrossRef |
Gscholar
(2)
Armenise L, Simeone MC, Piredda R, Schirone B (2012)Validation of DNA barcoding as an efficient tool for taxon identification and detection of species diversity in Italian conifers. European Journal of Forest Research 131: 1337-1353.
CrossRef |
Gscholar
(3)
Borsch T, Quandt D (2009)Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Systematics and Evolution 282: 169-199.
CrossRef |
Gscholar
(4)
Celinski K, Kijak H, Wojnicka-Póltorak A, Buczkowska-Chmielewska K, Sokolowska J, Chudzinska E (2017)Effectiveness of the DNA barcoding approach for closely related conifers discrimination: a case study of the
Pinus mugo complex. Comptes Rendus Biologies 340: 339-348.
CrossRef |
Gscholar
(5)
Darriba D, Taboada GL, Doallo R, Posada D (2012)jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772-772.
CrossRef |
Gscholar
(6)
De Vere N, Rich TCG, Ford CR, Trinder SA, Long C, Moore CW, Sattertwaite D, Davies H, Allainguillaume J, Ronca S, Tatarinova T, Garbett H, Walker K, Wilkinson MJ (2012)DNA Barcoding the Native Flowering Plants and Conifers of Wales. PLoS One 7: e37945.
CrossRef |
Gscholar
(7)
Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S (2015)ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports 5 (1): 313.
CrossRef |
Gscholar
(8)
Eckenwalder JE (2009)Conifers of the world: the complete reference. Timber Press, Portland, OR, USA, pp. 720.
Gscholar
(9)
Eckert AJ, Hall BD (2006)Phylogeny, historical biogeography, and patterns of diversification for
Pinus (Pinaceae): phylogenetic tests of fossil-based hypotheses. Molecular Phylogeny and Evolution 40: 166-182.
CrossRef |
Gscholar
(10)
Farjon A, Filer D (2013)An atlas of the world’s conifers: an analysis of their distribution, biogeography, diversity and conservation status. Brill, Leiden, Netherlands, pp. 512.
Gscholar
(11)
Gallien L, Saladin B, Boucher FC, Richardson DM, Zimmermann NE (2016)Does the legacy of historical biogeography shape current invasiveness in pines? New Phytologist 209: 1096-105.
CrossRef |
Gscholar
(12)
Gaussen H (1960)Les Gymnospermes actuelles et fossiles. Fascicule VI Chapitre XI, Généralités, genre
Pinus [Current and fossil Gymnosperms. Fascicle VI Chapter XI, General, genus
Pinus]. Travaux du Laboratoire Forestier de Toulouse, Faculté des Sciences, Toulouse, France. [in French]
Gscholar
(13)
Geada López G, Kamiya K, Harada K (2002)Phylogenetic relationships of
Diploxylon pines (Subgenus
Pinus) based on plastid sequence data. International Journal of Plant Sciences 163: 737-747.
CrossRef |
Gscholar
(14)
Gernandt DS, López G, García S, Liston A (2005)Phylogeny and classification of
Pinus. Taxon 4: 29-42.
CrossRef |
Gscholar
(15)
Gernandt DS, Magallón S, López G, Flores OZ, Willyard A, Liston A (2008)Use of simultaneous analyses to guide fossil-based calibrations of Pinaceae phylogeny. International Journal of Plant Sciences 169: 1086-1099.
CrossRef |
Gscholar
(16)
Gernandt DS, Aguirre X, Vázquez-Lobo A, Willyard A, Moreno A, Pérez De la Rosa JA, Piñero D, Liston A (2018)Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. American Journal of Botany 105: 1-15.
CrossRef |
Gscholar
(17)
Grotkopp E, Rejmánek M, Danderson MJ, Rost TL (2004)Evolution of genome size in pines (
Pinus) and its life-history correlates: supertree analyses. Evolution 58: 1705-1729.
CrossRef |
Gscholar
(18)
Hao DC, Chen SL, Xiao PG (2010)Sequence characteristics and divergent evolution of the chloroplast psbA-trnH noncoding region in gymnosperms. Journal of Applied Genetics 51: 259-273.
CrossRef |
Gscholar
(19)
Hernández-León S, Gernandt D, Pérez De la Rosa J, Jardón-Barbolla L (2013)Phylogenetic relationships and species delineation in
Pinus Section
Trifoliae inferred from plastid DNA. PLoS One 8 (7): e70501.
CrossRef |
Gscholar
(20)
Heuertz M, Teufel J, González-Martínez SC, Soto A, Fady B, Alía R, Vendramin GG (2010)Geography determines genetic relationships between species of mountain pine (
Pinus mugo complex) in Western Europe. Journal of Biogeography 37: 541-556.
CrossRef |
Gscholar
(21)
Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim KJ, Kress WJ, Schneider H, van Alphenstahl J, Barrett SC, van den Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TA, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim YD, Lahaye R, Lee HL, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park CW, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi DK, Little DP (2009a)A DNA barcode for land plants. Proceedings of the National Academy of Sciences USA 106 (31): 12794-12797.
CrossRef |
Gscholar
(22)
Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, Cowan R, Chase MW, Gaudeul M, Hollingsworth PM (2009b)Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources 9: 439-457.
CrossRef |
Gscholar
(23)
Hong Y-P, Krupkin AB, Strauss SH (1993)Chloroplast DNA transgresses species boundaries and evolves at variable rates in the Califormia close-cone pines (
Pinus radiata, P. muricata and
P. attenuata). Molecular Phylogeny and Evolution 2 (4): 322-329.
CrossRef |
Gscholar
(24)
Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS (2017)ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14 (6): 587-589.
CrossRef |
Gscholar
(25)
Karalamangala RR, Nickrent DL (1989)An electrophoretic study of representatives of subgenus
Diploxylon of
Pinus. Canadian Journal of Botany 67: 1750-1759.
CrossRef |
Gscholar
(26)
Kress WJ, Erickson DL (2007)A two-locus global DNA barcode for land plants: the coding
rbcL gene complements the non-coding
trnH-psbA spacer region. PLoS One 2 (6): e508.
CrossRef |
Gscholar
(27)
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005)Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences USA 102 (23): 8369-8374.
CrossRef |
Gscholar
(28)
Krupkin AB, Liston A, Strauss SH (1996)Phylogenetic analysis of the hard pines (
Pinus subgenus
Pinus, Pinaceae) from chloroplast DNA restriction site analysis. American Journal of Botany 83: 489-498.
CrossRef |
Gscholar
(29)
Lidholm J, Szmidt A, Gustafsson P (1991)Duplication of the
psbA gene in the chloroplast genome of two
Pinus species. Molecular and General Genetics 226: 345-352.
CrossRef |
Gscholar
(30)
Lidholm J, Gustafsson P (1991)A three-step model for the rearrangement of the chloroplast
trnK-psbA region of the gymnosperm
Pinus contorta. Nucleic Acids Research 19 (11): 2881-2887.
CrossRef |
Gscholar
(31)
Liston A, Robinson WA, Piñero D, Alvarez-Buylla ER (1999)Phylogenetics of
Pinus (Pinaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Molecular Phylogeny and Evolution 11: 95-109.
CrossRef |
Gscholar
(32)
Liston A, Parker-Defeniks M, Syring JV, Willyard A, Cronn R (2007)Interspecific phylogenetic analysis enhances intraspecific phylogeographical inference: a case study in
Pinus lambertiana. Molecular Ecology 16: 3926-3937.
CrossRef |
Gscholar
(33)
Matos JA, Schaal BA (2000)Chloroplast evolution in the
Pinus montezumae complex: a coalescent approach to hybridization. Evolution 54: 1218-1233.
CrossRef |
Gscholar
(34)
Müller KF, Quandt D, Müller J, Neinhuis C (2005)PhyDE
® 0.995: phylogenetic data editor. Web site.
Online |
Gscholar
(35)
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015)IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268-274.
CrossRef |
Gscholar
(36)
Olsson S, Grivet D, Cattonaro F, Vendramin V, Giovannelli G, Scotti-Saintagne C, Vendramin GG, Fady B (2020)Evolutionary relevance of lineages in the European black pine (
Pinus nigra) in the transcriptomic era. Tree Genetics and Genomes 16 (2): 508.
CrossRef |
Gscholar
(37)
Palmé AE, Pyhäjärvi T, Wachowiak W, Savolainen O (2009)Selection on nuclear genes in a
Pinus phylogeny. Molecular Biology and Evolution 26: 893-905.
CrossRef |
Gscholar
(38)
Parks M, Cronn R, Liston A (2009)Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7 (1): 19363.
CrossRef |
Gscholar
(39)
Parks M, Liston A, Cronn R (2011)Newly developed primers for complete
ycf1 amplification in
Pinus (Pinaceae) chloroplasts with possible family-wide utility. American Journal of Botany 98: 185-188.
CrossRef |
Gscholar
(40)
Parks M, Cronn R, Liston A (2012)Separating the wheat from the chaff: mitigating the effects of noise in a plastome phylogenomic data set from
Pinus L. (Pinaceae). BMC Evolutionary Biology 12 (1): 100.
CrossRef |
Gscholar
(41)
Price RA, Liston A, Strauss SH (1998)Phylogeny and systematics of
Pinus. In: “Ecology and Biogeography of
Pinus” (Richarson DM ed). Cambridge University Press, Cambridge, UK, pp. 49-68.
Online |
Gscholar
(42)
Rambaut A, Suchard MA, Xie Drummond D AJ (2014)Tracer v1.6. Web site.
Online |
Gscholar
(43)
Ran JH, Wang PP, Zhao HJ, Wang XQ (2010)A test of seven candidate barcode regions from the plastome in
Picea (Pinaceae). Journal of Integrative Plant Biology 52 (12): 1109-1126.
CrossRef |
Gscholar
(44)
Rieseberg LH, Soltis DE (1991)Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5 (1): 65-84.
Online |
Gscholar
(45)
Ronquist F, Teslenko M, Van Der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012)MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539-542.
CrossRef |
Gscholar
(46)
Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, Chaudhary LB, Datt B, Bag SK, Singh PK, Nair NK, Husain T, Tuli R (2010)Universal plant DNA barcode loci may not work in complex groups: a case study with Indian
Berberis species. PLoS One 5 (10): e13674.
CrossRef |
Gscholar
(47)
Saladin B, Leslie AB, Wüest RO, Litsios G, Conti E, Salamin N, Zimmermann NE (2017)Fossils matter: improved estimates of divergence times in
Pinus reveal older diversification. BMC Evolutionary Biology 17 (1): 49.
CrossRef |
Gscholar
(48)
Sass C, Little DP, Stevenson DW, Specht CD (2007)DNA barcoding in the Cycadales: testing the potential of proposed barcoding markers for species identification of Cycads. PLoS One 11: e1154.
CrossRef |
Gscholar
(49)
Scotti-Saintagne C, Giovannelli G, Scotti I, Roig A, Spanu I, Vendramin GG, Guibal F, Fady B (2019)Recent, late Pleistocene fragmentation shaped the phylogeographic structure of the European black pine (
Pinus nigra Arnold). Tree Genetics and Genomes 15 (5): 355.
CrossRef |
Gscholar
(50)
Strauss SH, Doerksen AH (1990)Restriction fragment analysis of pine phylogeny. Evolution 4: 1081-1096.
CrossRef |
Gscholar
(51)
Stöver BC, Müller KF (2010)TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7.
CrossRef |
Gscholar
(52)
Syring J, Willyard A, Cronn R, Liston A (2005)Evolutionary relationships among
Pinus (Pinaceae) subsections inferred from multiple low-copy nuclear loci. American Journal of Botany 92: 2086-2100.
CrossRef |
Gscholar
(53)
Tiffney BH (1985)The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. Journal of the Arnold Arboretum 66: 243-273.
CrossRef |
Gscholar
(54)
Vasilyeva G, Goroshkevich S (2019)Artificial crosses and hybridization frequency in five-needle pines. Dendrobiology 80: 123-130.
CrossRef |
Gscholar
(55)
Von Cräutlein M, Korpelainen H, Pietiläinen M, Rikkinen J (2011)DNA barcoding: a tool for improved taxon identification and detection of species diversity. Biodiversity and Conservation 20: 373-389.
CrossRef |
Gscholar
(56)
Von Raab-Straube E (2014)Gymnospermae. In: “Euro+Med Plantbase - The Information Resource For Euro-mediterranean Plant Diversity”. Web site.
Online |
Gscholar
(57)
Wang XR, Tsumura Y, Yoshimaru H, Nagasaka K, Szmidt AE (1999)Phylogenetic relationships of Eurasian pines (
Pinus, Pinaceae) based on chloroplast
rbcL, MATK, RPL20-RPS18 spacer, and
TRNV intron sequences. American Journal of Botany 86 (12): 1742-1753.
CrossRef |
Gscholar
(58)
Whittemore AT, Schaal BA (1991)Interspecific gene flow in sympatric oaks. Proceedings of the National Academy of Sciences USA 88 (6): 2540-2544.
CrossRef |
Gscholar
(59)
Willyard A, Cronn R, Liston A (2009)Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Molecular Phylogeny and Evolution 52: 498-511.
CrossRef |
Gscholar
(60)
Yan M, Xiong Y, Liu R, Deng M, Song J (2018)The application and limitation of universal chloroplast markers in discriminating East Asian evergreen oaks. Frontiers in Plant Science 9: 235.
CrossRef |
Gscholar