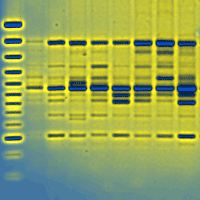
Retrotransposable elements are important and peculiar genetic components derived from ancient retrovirus insertion inside plants genome. Their ability to move and/or replicate inside the genome is an important evolutionary force, responsible for the increase of genome size and the regulation of gene expression. Retrotransposable elements are well characterized in model or crop species like Arabidopsis thaliana and Oryza sativa, but are poorly known in forest tree species. In this paper the molecular identification of retrotransposable elements in Fagus sylvatica L. is reported. Two retrotransposons, belonging to the two major classes of LTR and non-LTR elements, were characterized trough a SCAR (Sequence Characterized Amplified Region) strategy. The analysis demonstrated the presence of multiple copies of retrotransposable elements inside the genome of beech, in accordance with the viral quasi-species theory of retrotransposon evolution. The cloning and sequencing of amplification products and a Cleaved Amplified Polymorphisms (CAPs) approach on the identified retrotransposons, showed a high level of diversity among the multiple copies of both elements. The identification of retrotransposable elements in forest trees represents an important step toward the understanding of mechanisms of genome evolution. Furthermore, the high polymorphism of retrotransposable elements can represent a starting point for the development of new genetic variability markers.
Keywords
, , ,
Citation
Emiliani G, Paffetti D, Giannini R (2009). Identification and molecular characterization of LTR and LINE retrotransposable elements in Fagus sylvatica L.. iForest 2: 119-126. - doi: 10.3832/ifor0501-002
Paper history
Received: Feb 04, 2009
Accepted: Mar 25, 2009
First online: Jun 10, 2009
Publication Date: Jun 10, 2009
Publication Time: 2.57 months
© SISEF - The Italian Society of Silviculture and Forest Ecology 2009
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.

Breakdown by View Type
(Waiting for server response...)
Article Usage
Total Article Views: 66413
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 55598
Abstract Page Views: 3942
PDF Downloads: 5774
Citation/Reference Downloads: 60
XML Downloads: 1039
Web Metrics
Days since publication: 6082
Overall contacts: 66413
Avg. contacts per week: 76.44
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2009): 3
Average cites per year: 0.18
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
(1)
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997)Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389-3402.
CrossRef |
Gscholar
(2)
Asins MJ, Monforte AJ, Mestre PF, Carbonel EA (1999)Citrus and
Prunus copia-like retrotransposons. Theoretical and Applied Genetics 99: 503-510.
CrossRef |
Gscholar
(3)
Biémont C, Vieira C (2005)What transposable elements tell us about genome organization and evolution: the case of
Drosophila. Cytogenetic and Genome Research 110: 25-34.
CrossRef |
Gscholar
(4)
Casacuberta JM, Santiago N (2003)Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311: 1-11.
CrossRef |
Gscholar
(5)
Casacuberta JM, Vernhettes S, Audeon C, Grandbastien MA (1997)Quasispecies in retrotransposons: a role for sequence variability in Tnt1 evolution. Genetica 100: 109-117.
CrossRef |
Gscholar
(6)
Craig NL, Craigie R, Gellert M, Lambowitz AM (2002)Mobile DNA II.American Society for Microbiology Press, Washington, DC.
Gscholar
(7)
Dewannieux M, Heidmann T (2005)LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling. Cytogenetic and Genome Research 110: 35-48.
CrossRef |
Gscholar
(8)
Durbin ML, Denton AL, Clegg MT (2001)Dynamics of mobile element activity in chalcone synthase loci in the common morning glory (
Ipomea purpurea). PNAS 98(9): 5084-5089.
CrossRef |
Gscholar
(9)
Emiliani G, Paffetti D, Vettori C, Giannini R (2004)Geographic distribution of genetic variability of
Fagus sylvatica L. in southern Italian populations. Forest Genetics 11: 231-237.
Gscholar
(10)
Feschotte C, Jiang N, Wessler SR (2002)Plant transposable elements: where genetics meets genomics. Nature Reviews Genetics 3: 329-341.
CrossRef |
Gscholar
(11)
Flowers JM, Purugganan MD (2008)The evolution of plant genomes scaling up from a population perspective. Current Opinion in Genetics & Development 18: 565-570.
CrossRef |
Gscholar
(12)
Friesen N, Brandes A, Heslop-Harrison JS (2001)Diversity, origin, and distribution of retrotransposons (
gypsy and
copia) in conifers. Molecular Biology and Evolution 18: 1176-1188.
Gscholar
(13)
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N et al. Jaillon O, et al. (2007)The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463-468.
CrossRef |
Gscholar
(14)
Kalendar R, Grob, Regina M, Suoniemi A, Schulman A (1999)IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theoretical and Applied Genetics 98: 704-711.
CrossRef |
Gscholar
(15)
Kalendar R, Schulman AH (2006)IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nature Protocols 1(5): 2478-2484.
CrossRef |
Gscholar
(16)
Kamm A, Doudrick RL, Heslop-Harrison JS, Schmidt T (1996)The genomic and physical organization of Ty1-copia-like sequences as a component of large genomes in
Pinus elliottii var.
elliottii and other gymnosperms. PNAS 93: 2708-2713.
CrossRef |
Gscholar
(17)
Kapitonov VV, Jurka J (2003)Molecular paleontology of transposable elements in the
Drosophila melanogaster genome. PNAS 100(11): 6569-6574.
CrossRef |
Gscholar
(18)
Kazazian HH (2004)Mobile elements: drivers of genome evolution.Science303: 1626-1632.
CrossRef |
Gscholar
(19)
L’Homme Y, Séguin A, Tremblay FM (2000)Different classes of retrotransposons in coniferous spruce species. Genome 43: 1084-1089.
CrossRef |
Gscholar
(20)
Larkin M, Blackshields G. Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007)ClustalW and ClustalX version 2. Bioinformatics 23(21): 2947-2948.
CrossRef |
Gscholar
(21)
Leeton PR, Smyth DR (1993)An abundant LINE-like element amplified in the genome of
Lilium speciosum. Molecular Genetics and Genomics 237: 97-104.
Gscholar
(22)
Leprince AS, Grandbastien MA, Meyer C (2001)Retrotransposons of the Tnt1B family are mobile in
Nicotiana plumbaginifolia and can induce alternative splicing of the host gene upon insertion. Plant Molecular Biology 47: 533-541.
CrossRef |
Gscholar
(23)
Meyers BC, Tingey SV, Morgante M (2001)Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Research 11: 1660-1676.
CrossRef |
Gscholar
(24)
Morgante M (2006)Plant genome organization and diversity: the year of the junk! Current Opinion in Biotechnology 17:168-173.
Gscholar
(25)
Nikaido M, Okada N (2000)CetSINEs and AREs are not SINEs but are parts of cetartiodactyl L1. Mammalian Genome 11: 1123-11236.
CrossRef |
Gscholar
(26)
Paffetti D, Scotti C, Gnocchi S, Fancelli S, Bazzicalupo M (1996)Genetic diversity of an Italian
Rhizobium meliloti population from different
Medicago sativa varieties. Applied Environmental Microbiology 62: 2279-2285.
Gscholar
(27)
Queen RA, Gribbon BM, James C, Jack P, Flavell AJ (2004)Retrotransposon based molecular markers for linkage and genetic diversity analysis in wheat. Molecular Genetics and Genomics 271: 91-97.
CrossRef |
Gscholar
(28)
Rozen S, Skaletsky HJ (2000)Primer3 on the WWW for general users and for biologist programmers. In: “Bioinformatics methods and protocols: methods in molecular biology” (Krawetz S, Misener S eds). Humana Press, Totowa, NJ, pp. 365-386.
Gscholar
(29)
SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996)Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765-768.
CrossRef |
Gscholar
(30)
Smykal P (2006)Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. Journal of Applied Genetics 47: 221-230.
Gscholar
(31)
Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR Grandbastien M-A (2005)Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theoretical and Applied Genetics 110: 819-831.
CrossRef |
Gscholar
(32)
Tamura K, Dudley J, Nei M, Kumar S (2007)MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596-1599.
CrossRef |
Gscholar
(33)
TIHGSC (2001)Initial sequencing and analysis of the human genome. The International Human Genome Sequencing Consortium. Nature 409 (6822): 860-921.
Gscholar
(34)
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, GrigorievI, Hellsten U, et al. (2006)The genome of black cottonwood,
Populus trichocarpa (Torr. and Gray). Science 313: 1596-1604.
CrossRef |
Gscholar
(35)
Vance V, Vaucheret H (2002)RNA silencing in plants - defense and counterdefense. Science 292: 2277-2280.
CrossRef |
Gscholar
(36)
Venturi S, Dondini L, Donini P, Sansavini (2006)Retrotransposons characterization and fingerprinting of apple clones by S-SAP markers. Theoretical and Applied Genetics 112: 440-444.
CrossRef |
Gscholar
(37)
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999)Retrotransposon BARE-1 and its role in genome evolution in the genus
Hordeum. Plant Cell 11: 1769-1784.
Gscholar
(38)
Vitte C, Bennetzen JL (2006)Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. PNAS 103(47): 17638-17643.
CrossRef |
Gscholar
(39)
Vitte C, Panaud O (2005)LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenetic and Genome Research 110: 91-107.
CrossRef |
Gscholar
(40)
Vitte C, Panaud O, Quesneville H (2007)LTR retrotransposons in rice (
Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics 8: 218.
CrossRef |
Gscholar
(41)
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W (1997)Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Molecular Genetics and Genomics 253: 687-694.
Gscholar
(42)
Wessler SR (1998)Transposable elements associated with normal plant genes. Physiologia Plantarum 103: 581-586.
CrossRef |
Gscholar
(43)
White SE, Habera LF, Wessler SR (1994)Retrotransposon in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. PNAS 91: 11792-11796.
CrossRef |
Gscholar
(44)
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007)A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics 8: 973-982.
CrossRef |
Gscholar
(45)
Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap1 E (2008)A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science319: 1527-1530.
CrossRef |
Gscholar
(46)
Yoshioka Y, Matsumoto S, Kojima S, Ohshima K, Okada N, Machida Y (1993)Molecular characterization of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific tRNAs. PNAS 90: 6562-6566.
CrossRef |
Gscholar