Molecular evidence of bidirectional introgression between Quercus suber and Quercus ilex
iForest - Biogeosciences and Forestry, Volume 11, Issue 2, Pages 338-343 (2018)
doi: https://doi.org/10.3832/ifor2570-011
Published: Apr 18, 2018 - Copyright © 2018 SISEF
Research Articles
Collection/Special Issue: INCOTW - Sassari, Italy (2017)
International Congress on Cork Oak Trees and Woodlands
Guest Editors: Piermaria Corona, Sandro Dettori
Abstract
Cork oak and holm oak share a large part of their natural range, and are known to hybridize in mixed stands. This hybridization is supposed to have played a relevant role in the past history of cork oak. Previous research has reported that F1 hybrids are produced with holm oak acting as pollen recipient, therefore carrying holm oak chloroplast. Additionally, F1 hybrids have been assumed to be pollinated mostly by cork oak. Continued backcrossing of F1 hybrids with cork oak (supported by flowering phenology) could have created the organellar introgression patterns observed nowadays in Eastern Spain and Southern France cork oak populations. On the contrary, no organellar introgression has been detected in holm oak and multiple generation backcross individuals to holm oak have not been reported so far. In this work, we examined whether hybrids preferentially backcross with cork oak or with holm oak. To reach this goal, we genotyped by using eight microsatellite loci the progeny of four cork and four holm oak trees (33 and 44 half-siblings, respectively), and of four hybrids (468 half-siblings) collected over three years from a natural mixed population. We used the STRUCTURE software to estimate the proportion of the genotype of each seedling inherited from cork oak (qs) or from holm oak (qi). The ratio of the offspring q value over the mother q value helped determine the source of pollen that originated each acorn. Our results show for the first time that hybrid trees can be effectively pollinated by both parental species. Additionally, each hybrid tree was predominantly pollinated by the most abundant oak species in its vicinity. These results confirm the occurrence of bidirectional introgression, previously suggested for adult hybrid trees in the field, and point out the pattern of introgression in the seedlings could be most affected by the abundance of the parental species.
Keywords
Cork Oak, Holm Oak, Hybridization, Introgression, Microsatellites
Introduction
Hybridization between cork oak (Quercus suber L.) and holm oak (Q. ilex L.) has been known for a long time. The first written references to putative hybrid individuals date back to the mid-19th century ([5], [21], [3]). The ranges of cork and holm oak overlap in the western Mediterranean basin, where they form mixed stands on acid and decarbonated soils, the preferred ones for cork oak. Morphologically intermediate, presumably hybrid individuals, can be found in these stands, and are easily identifiable by their bark, and by some leaf intermediate traits, such as leaf thickness, which show characteristics of both parental species ([21]). These putative hybrid individuals occur at low frequency and are scattered among the pure cork and holm oaks. Introgression of genes from one species to another can have important adaptive and evolutionary consequences ([13], [30]). In the case of holm and cork oaks, it has been proposed that such process may have helped cork oak thrive in adverse environmental conditions (e.g., during the glacial pulses of the Pleistocene - [24]).
Molecular markers have confirmed the hybrid nature of phenotypically intermediate individuals and were used to assess the extent of hybridization between Q. suber and Q. ilex in natural mixed populations. Analysis of chloroplast DNA (cpDNA), which is maternally inherited in oaks, detected Q. suber populations in Southern France and Eastern Spain with a typical Q. ilex lineage cpDNA ([15], [23], [25]). Phenology of both species as well as post pollination barriers as those reported by Boavida et al. ([2]) favor hybridization with Q. suber acting as pollen donor, so that hybrids carry the ilex chloroplast. Chloroplast introgression pattern suggests hybrid trees carrying the ilex chloroplast recurrently backcrossed with Q. suber as pollen donor. Using isozymes ([11], [28]) or nuclear microsatellites ([34], [4]), previous studies confirmed the hybrid character of morphologically intermediate oak trees, and showed the current existence of hybridization. Burgarella et al. ([4]) estimated a low rate of ongoing introgression in cork oak over its range (~2%), a result confirmed by Lumaret & Jabbour-Zahab ([26] - ~4%). These studies suggested that introgression could occur bidirectionally. However, if the flowering phenology of first generation hybrids (F1 hybrids) is more similar to Q. suber, as suggested by Perea García-Calvo ([29]), they would preferentially backcross with Q. suber, while mating with Q. ilex, especially when the latter species acts as pollen donor, would be much less frequent.
In the current work, we genotyped seedlings of morphologically putative hybrid trees, to identify backcrossing events with either Q. ilex or Q. suber as pollen donors, and to quantify the frequency of introgression towards each of the parental species. Our study provides a deeper insight in the hybridization and introgression processes which could be currently taking place between these two species.
Material and methods
Sampling site and acorn collection
The sampling site is located in Central Spain, (39° 59′ N, 05° 07′ W) in a mixed “dehesa”, i.e., highly anthropized open woodland. Quercus suber and Q. ilex are the dominant species, while there are some stands with comparatively few Q. faginea individuals. Four mature hybrid trees were identified by their phenotypes. These hybrid trees occurred in three distinct areas (IS, LG, and ZL) which differed in tree density and species composition. One hybrid was identified in IS, where Q. ilex and Q. suber occur at a density of 38 trees ha-1 and where Q. suber is slightly more abundant. Two hybrids were found in LG, approximately 25 m apart and 740 m away from the IS hybrid. At LG, tree density was low (20 trees ha-1) and dominated by Q. ilex. The fourth hybrid tree was found in ZL, where tree density was 123.5 trees ha-1 , with more Q. suber trees (38.3 trees ha-1 for Q. ilex and 85.2 trees ha-1 for Q. suber), and approximately 2780 m away from the two LG hybrids and 3140 m away from the IS hybrid.
Acorns were collected in 2011, 2012 and 2014 on each of the four hybrids trees and also from four randomly selected cork oak and four holm oak trees located in IS (Tab. 1). Each year, all acorns from LG2 and ZL hybrid trees were collected, as well as approximately 1/3 of the acorns on the LG1 and IS hybrids.
Tab. 1 - Number of acorns sewed, germinated, and genotyped (sewed-germinated-genotyped) categorized by mother tree (family), taxa and cohort.
| Taxa | Genotype | 2011 | 2012 | 2014 |
|---|---|---|---|---|
| Hybrids | LG1 | 184 - 155 - 148 | 36 - 24 - 21 | 59 - 39 - 38 |
| LG2 | 19 - 3 - 3 | - | 14 - 5 - 5 | |
| IS | 106 - 85 - 84 | 76 - 61 - 59 | 95 - 77 - 77 | |
| ZL | 12 - 0 - 0 | 36 - 24 - 19 | 34 -14 - 14 | |
| Total | 321- 243 - 235 | 148 - 109 - 99 | 202 - 135 - 134 | |
| Q. ilex | I1 | - | 22 - 21 - 19 | 5 - 5 - 5 |
| I2 | - | 12 - 11 - 9 | 5 - 4 - 4 | |
| I3 | - | - | 4 - 4 - 4 | |
| I4 | - | - | 5 - 3 - 3 | |
| Total | - | 34 - 32 - 28 | 19 - 16 - 16 | |
| Q. suber | S1 | - | 12 - 8 - 8 | 4 - 2 - 2 |
| S2 | - | 18 - 12 - 11 | 5 - 4 - 4 | |
| S3 | - | - | 5 - 4 - 4 | |
| S4 | - | - | 5 - 4 - 4 | |
| Total | - | 30 - 20 - 19 | 19 - 14 - 14 |
All collected acorns were sown in peat-perlite substrate (3:1 proportion) in 3L containers in a greenhouse room kept at 22 °C, with a relative humidity of 50-60%. Seedlings were watered daily for 8 min. and five months after sowing, germination rate was estimated and leaf tissue collected from the surviving seedlings for DNA extraction.
Genotyping
DNA extraction was performed with a modification of the protocol by Doyle & Doyle ([8], [9]). We used eight polymorphic nuclear microsatellites (nSSRs) for genotyping: MSQ4, MSQ13 ([7]), QpZAG9, QpZAG15, QpZAG36, ([37]), QrZAG7, QrZAG11 and QrZAG20 ([18]). Microsatellites were amplified under standard PCR conditions ([34], [35]). Fluorescence labelled PCR products were analyzed in a 4300 LI-COR® automated sequencer (LI-COR Biosciences, Lincoln, NE, USA). Allele sizes were determined with the SAGA Microsatellite Analysis Software (LI-COR Biosciences, Lincoln, NE, USA). The presence of null alleles was examined using ML-Null ([17]).
Nuclear admixture analyses
We carried out admixture analyses using the Bayesian clustering approach implemented in the program STRUCTURE ver. 2.3.4 ([32]). In this model, the posterior probability (q) provides an estimation of the proportion of an individual genome originating from each of K differentiated genetic pools. Values of K ranging from 1 to 5 were tested in order to find the optimal K value. A value of K=2 was identified as the optimal K value following the methodology described by Evanno et al. ([12]) and was used in all subsequent analyses. In order to improve q estimations ([4]), we included in the STRUCTURE analyses the genotypes of approximately 95 Q. ilex and 96 Q. suber parental trees from the same stand, previously analyzed by Soto et al. ([35]), along with 59.671 virtual individuals simulated with the software SimHyb ([36]). These simulated individuals included a majority of parental trees with some F1 hybrids and backcrosses (Tab. S1 in Supplementary material).
Calculations were carried out under the admixture model assuming independent allele frequencies, as it has been done previously ([4]). A burn-in of 50.000 steps followed by 100.000 iterations were used, after verifying that results did not vary significantly across runs or with longer burn-in/iteration cycles. Assignation of seedlings to specific categories based on the qs values (estimation of the proportion of the genome coming from Q. suber) used the following thresholds: qs ≥ 0.85: suber; 0.85 < qs ≤ 0.65: suberoid; 0.65 ≤ qs < 0.35: hybrid; 0.35 ≤ qs < 0.15: ilicioid; qs ≤ 0.15: ilex. The software Colony ([16]) was used to infer selfing rates in the progeny using the adult trees as potential fathers and allowing up to one mismatch per seedling, and to detect potential crosses between hybrid trees.
The software R ([33]) was used to draw graphics, linear regressions between putative paternal qs and qso/qsm (the ratio between the offspring and the maternal qs), and to prepare a script to obtain all the possible paternal contributions for each offspring.
Results and discussion
Germination rates
A total of 671 acorns were collected from hybrid trees, and 53 and 49 acorns from Q. ilex and Q. suber, respectively (Tab. 1). Hybrid trees from LG2 and ZL produced fewer acorns than hybrid trees from LG1 and IS, and their germination rates were also lower (24% and 46%, respectively), while germination rates were above 70% for hybrids from LG1 and IS, as well as for the parental species trees (Fig. S1 in Supplementary material).
Assignation to specific categories
We did not detect null alleles for any locus. Autofecundation was ruled out, as the estimated selfing probability was <0.001 for every hybrid seedling. Moreover, mating between hybrid trees was also discarded after inspection of potential parents for each offspring estimated by Colony. The most probable number of differentiated genetic pools in the population was K = 2 (Fig. S2). The qs values (estimation of the proportion of the genome coming from Q. suber) obtained for the hybrid trees were 0.8763 for IS, 0.5988 for LG1, 0.8680 for LG2 and 0.5489 for ZL. These results would classify the hybrid trees at LG1 and ZL as most likely first generation hybrids (F1), and hybrid trees at IS and LG2 as more “advanced introgressed” individuals, at least second-generation backcrosses with Q. suber. These results are consistent with preferential backcrossing with Q. suber.
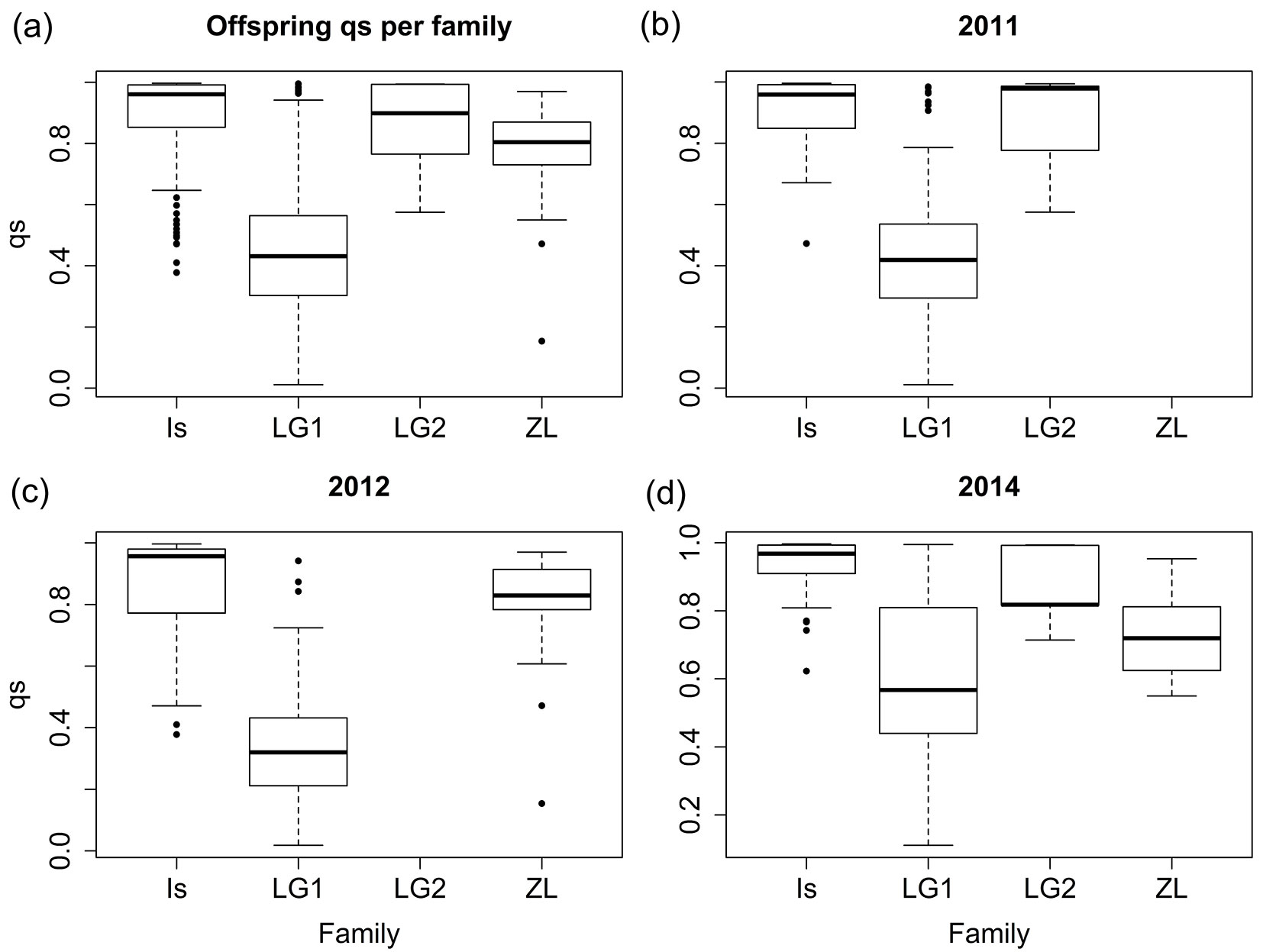
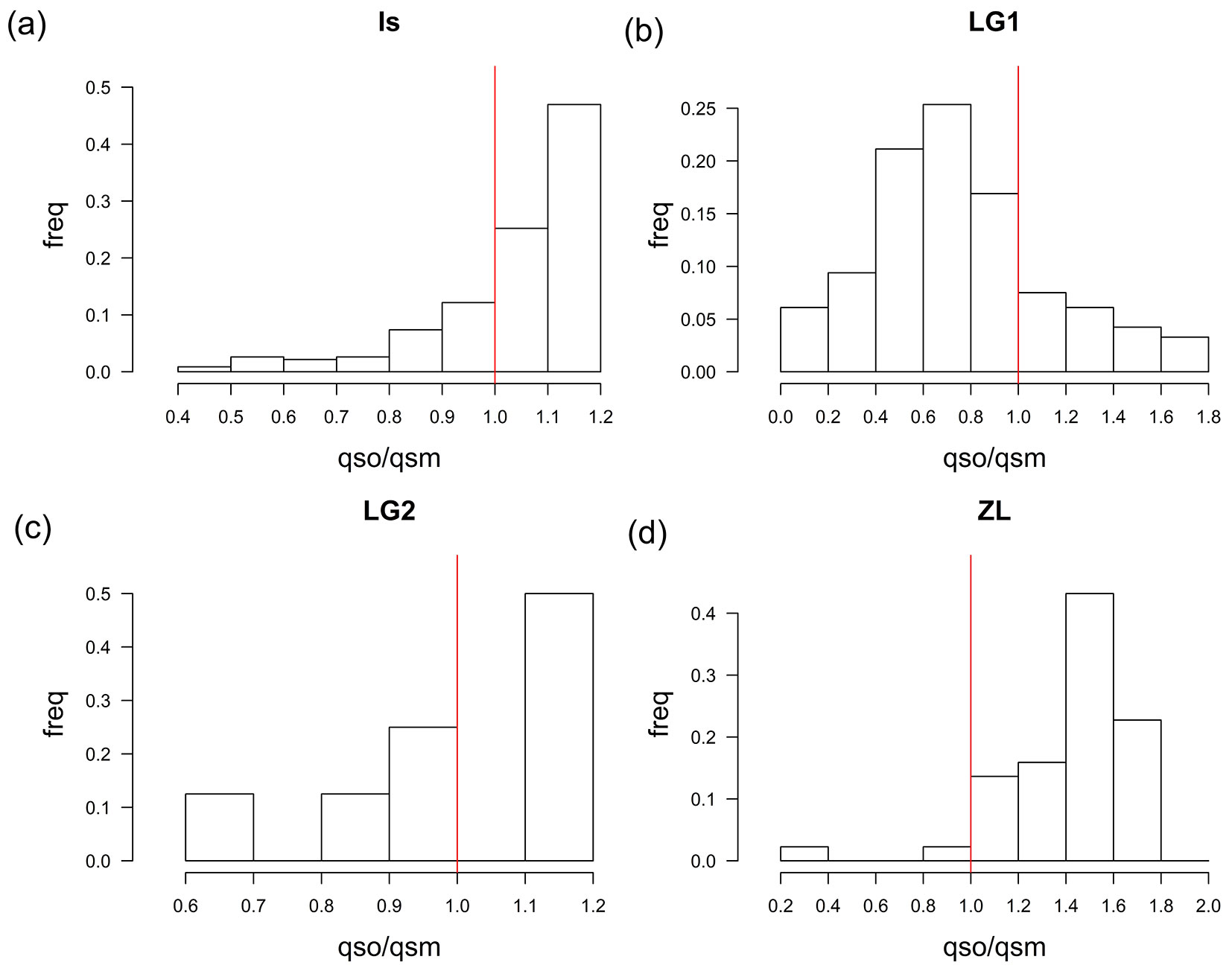
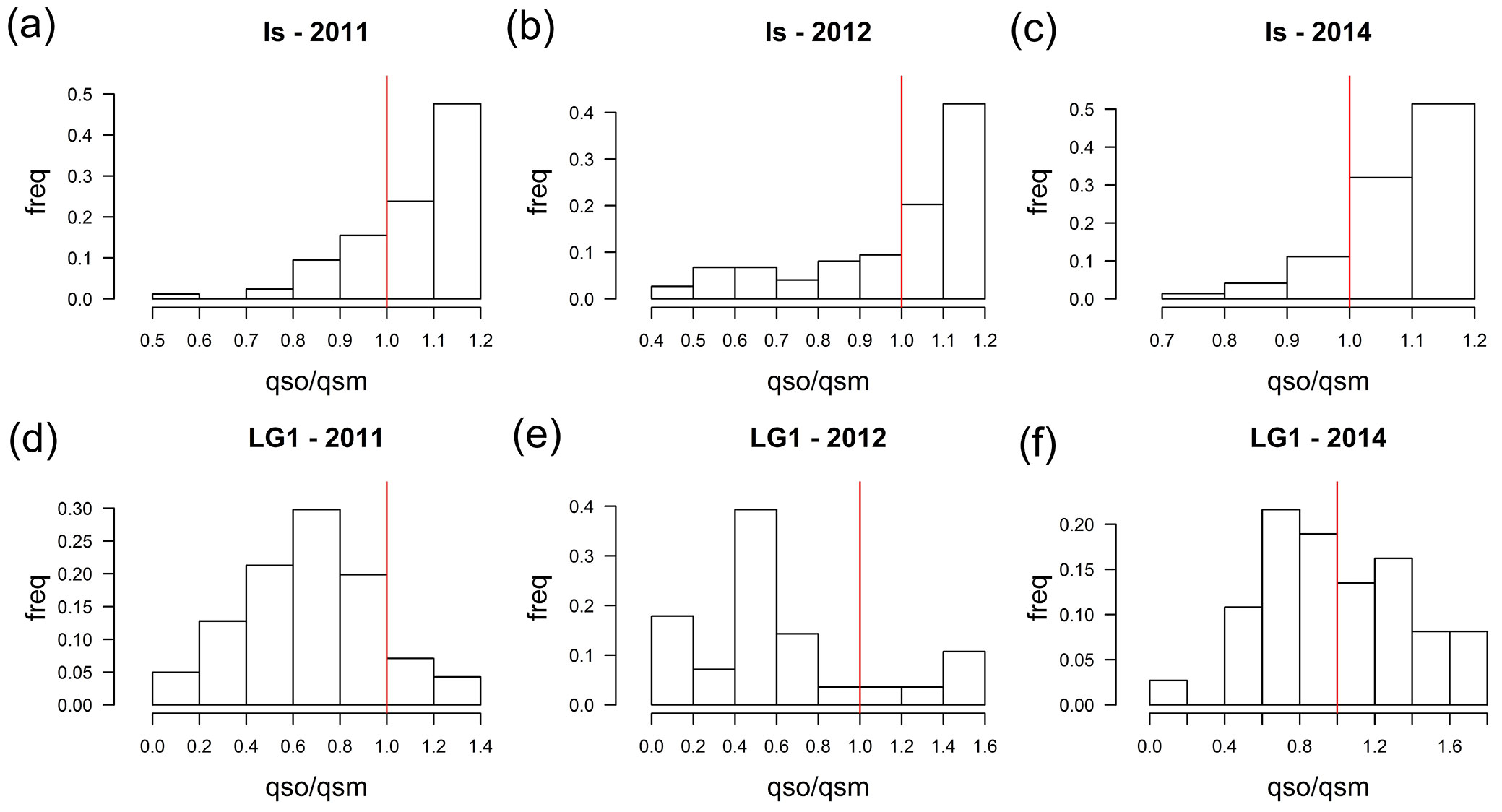
Then, qs values were also estimated for the progenies of the hybrids (Fig. 1). The high qs values obtained for most of the hybrid offspring, particularly for IS, LG2 and ZL, indicate a high contribution of Q. suber to their genomes and therefore previous works could have classified these seedlings as backcrosses to Q. suber. Conversely, the offspring of LG1 shows a broader range of qs, with an average value of 0.4512, closer to Q. ilex. However, assignation to specific classes based exclusively on qs may be biased using this set of markers, especially for advanced introgressed individuals ([36]). Actually, more than 30% of the virtual introgressed individuals included in the simulation as control would be misassigned according to qs. Following Soto et al. ([36]), we used the ratio qso/qsm to determine the most likely genetic pool of the pollen donor for each offspring (Fig. 2). These data support backcrossing with both parental species with many of the LG1 offspring having Q. ilex as father, and many offspring of the IS and ZL hybrid trees having Q. suber as father. Offspring of the LG2 hybrid tree had either Q. suber or Q. ilex as father. Fig. 3 shows the distribution of the qs ratio throughout the studied years for IS and LG1, the trees that produced more acorns. Although the general trends are kept for the whole period, some differences among years can be noticed, particularly for LG1: in 2011, probably a masting year with high acorn production, the vast majority of the seedlings came from Q. ilex pollen (84.5%), while in 2014, with a much lower acorn production, the proportion of effective pollinations from Q. ilex decreased to 54.1%.
Fig. 1 - Boxplots of the qs parameter across families for (a) all years combined and separately for (b) 2011, (c) 2012 , and (d) 2014. Sample sizes are presented in Tab. 1.
Fig. 2 - Histograms of the relative frequencies of the qso/qsm ratio between the offspring and the maternal qs for Is (a), LG1 (b), LG2 (c), and ZL (d) families. The bars to the right of the vertical red line indicate pollination by individuals with a higher percentage of suber genomes. The bars to the left of the vertical red line indicate pollination by individuals with a higher percentage of ilex genomes.
Fig. 3 - Histograms for the relative frequencies of the ratio between the offspring and the maternal qs for the families Is in 2011 (a), 2012 (b), 2014 (c), and LG1 in 2011 (d), 2012 (e), and 2014 (f). The bars to the right of the vertical red line indicate pollination by individuals with a higher percentage of suber genomes. The bars to the left of the vertical red line indicate pollination by individuals with a higher percentage of ilex genomes.
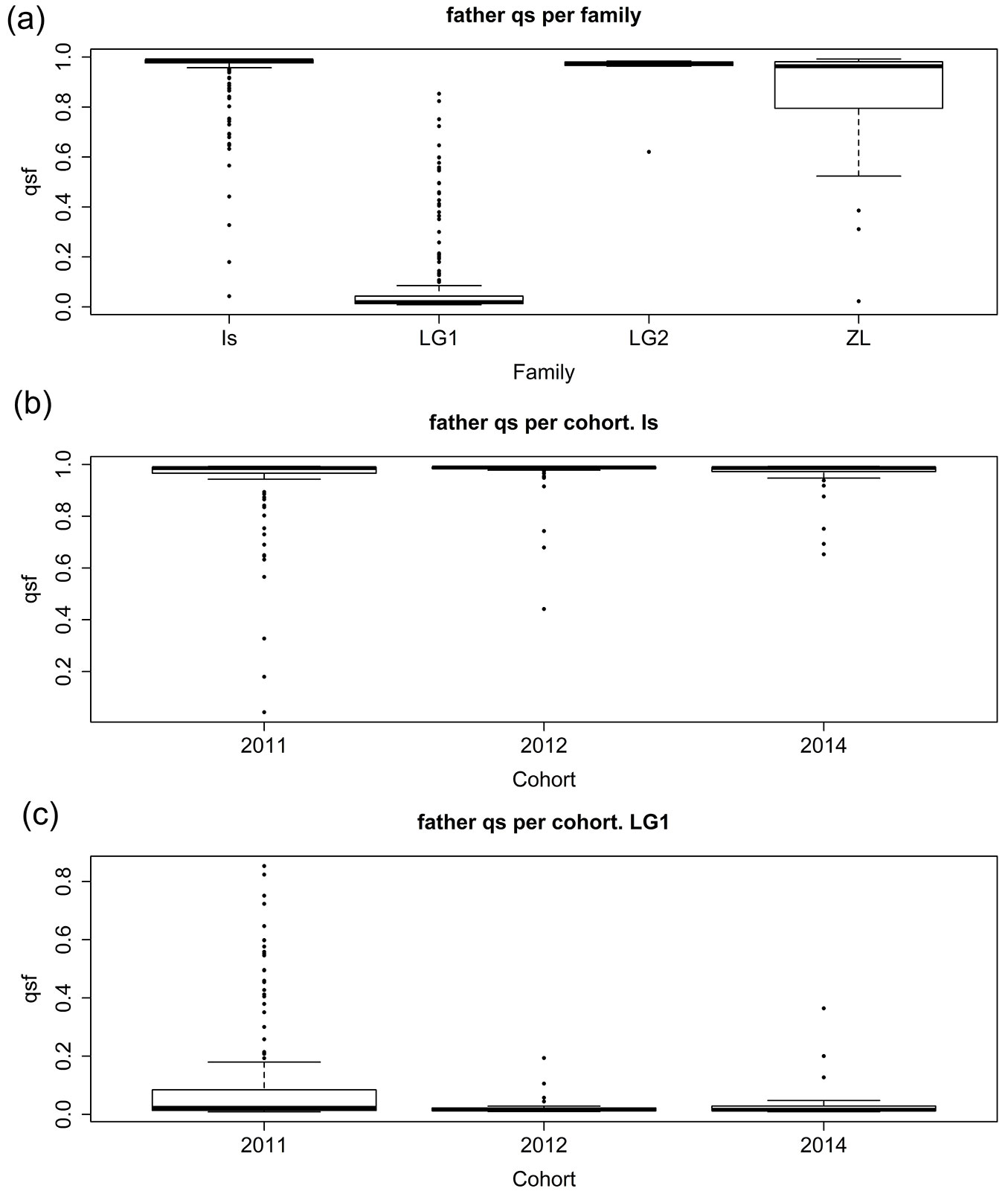
To get additional support for the results on pollination direction we prepared an R script to compare each individual offspring and its mother’s genotypes, and to obtain all the possible paternal genetic contributions (haplotypes). We then estimated their qs value using STRUCTURE. Fig. 4a shows a boxplot of the distribution of the qs values assigned to all the possible paternal haplotypes for each mother tree, and Fig. 4b and Fig. 4c show these distribution through the studied years for IS and LG1.
Fig. 4 - Boxplots of the qs parameter of the inferred putative fathers across families (a), and across cohorts for Is (b) and LG1 (c) families.
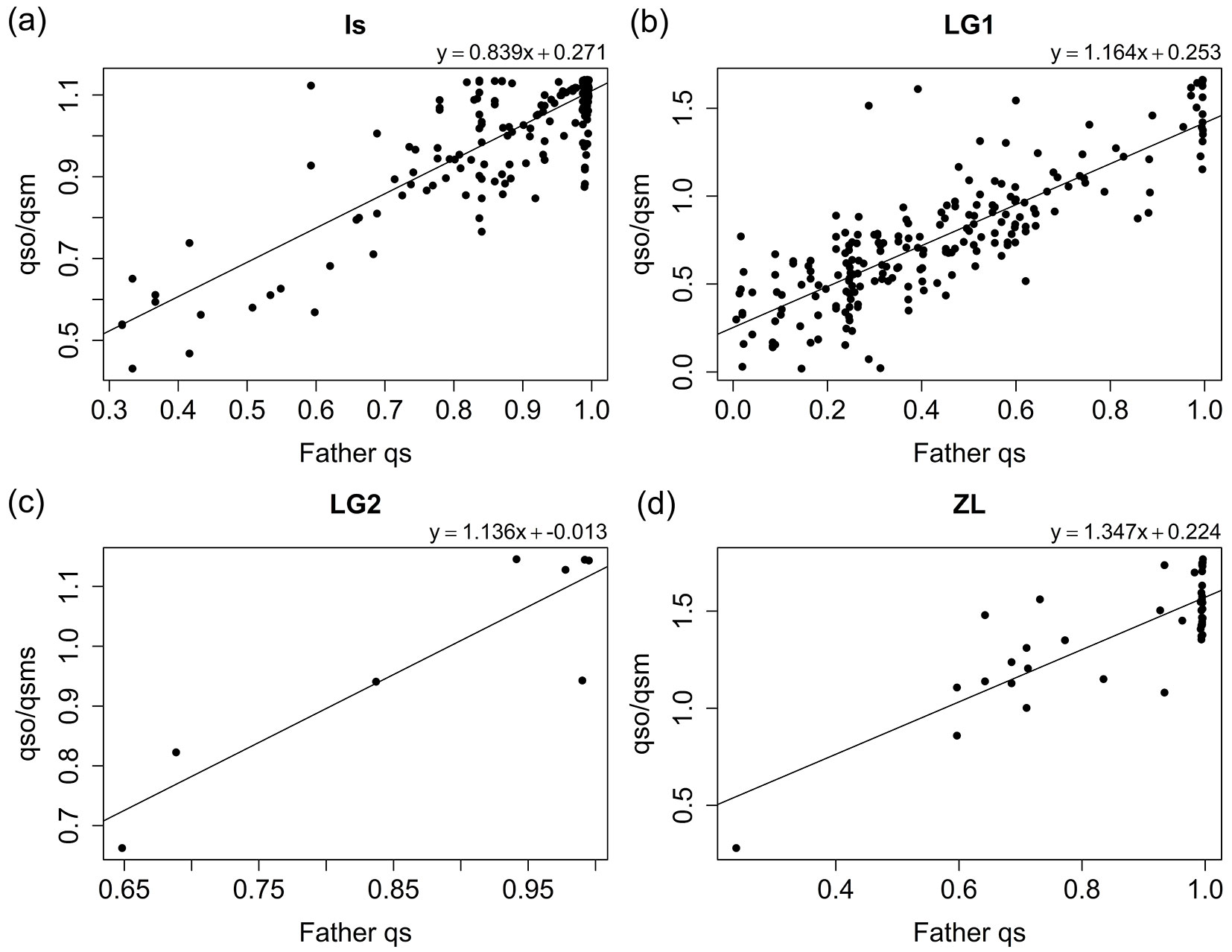
Finally, Fig. 5 shows the significant correlations above 0.80 for the estimated paternal qs and the ratio qso/qsm, especially for IS (Fig. 5a) and LG1 (Fig. 5b), the trees with larger progenies. This result confirms that using the ratio qso/qsm is a good approach to identify the genetic pool of the pollen donor, provided the mother tree is known.
Fig. 5 - Correlation of the ratio between the offspring and the maternal qs and the qs parameter of the inferred putative fathers for Is (a), LG1 (b), LG2 (c), and ZL (d) families. Fitted linear model for each family are indicated.
These results support bidirectional introgression, where hybrid trees backcross with both parental species. Bidirectional introgression was also suggested by genotypic data collected by Burgarella et al. ([4]) using the same set of nuclear microsatellite markers, on adult trees phenotypically classified as Q. suber and as Q. ilex sampled all across the overlapping distribution ranges of these two species. Lumaret & Jabbour-Zahab ([26]), using the same set of microsatellite markers, also reported the possibility of bidirectional introgression.
Each adult hybrid receives pollen from both parental species but is preferentially pollinated by either Q. ilex or Q. suber (Fig. 2). The major pollen donor for each hybrid tree seems to reflect the abundance of parental tree species in the area. For example, the hybrid tree LG1, located in La Laguna, is mostly pollinated by Q. ilex, the dominant species in that area (the number of seedlings from LG2 is too low to be conclusive); on the other hand, IS and ZL hybrids produced more seedlings from Q. suber pollen, the more abundant species in these areas. Therefore, the frequency of backcrossing with a parental species could be at least partially driven by pollen availability. Simulation studies using the oak complex Q. robur × Q. petraea have shown that the direction of introgression can be influenced by the composition of the pollen pool on individuals or stigmas which is itself affected by the spatial configuration of the parental species ([20], [19]). The direction of introgression depending on the abundance of the parental species in an area would be consistent with the findings in Eucalyptus ([14]), Senecio ([31]) and some mangroves ([39]).
Based on differential growth rate of pollen tubes, Boavida et al. ([2]) reported significantly higher success rate for the interspecific crosses with Q. suber acting as pollen donor rather than as female parent. However, the high number of hybrid offspring having Q. ilex as pollen donor, supports at least the partial breakage of this prezygotic incompatibility between introgressed hybrids and Q. ilex pollen. Notwithstanding, a potential effect of post-zygotic barriers on the observed gene flow directionality cannot be discarded. If some type of incompatibility existed between the two parental species, a high proportion of viable embryos could be expected for families of parent trees that have similar specific categories (estimated in our case through qs). Interestingly, the majority of seedlings of the IS parent, classified as an “advanced introgressed” cork oak tree (qs = 0.8763), had Q. suber as pollen donor. However, although both ZL and LG1 hybrids show intermediate qs values (0.5489 and 0.5988, respectively), they show contrasting proportions of effective pollinations from Q. suber and Q. ilex. As previously stated, the Q. ilex neighborhood of LG1 could account for the abundance of LG1 × Q. ilex backcrosses. In the case of ZL an overabundance of backcrosses to Q. suber was scored, considering the specific composition of the surrounding trees. Although the number of seedlings from this tree is still low to draw definitive conclusions, this fact could be due to a phenological synchrony between this tree’s female flowers and pollination of the surrounding Q. suber ([29]), or to post-pollination processes ([38]), such as genomic incompatibilities between this hybrid and surrounding Q. ilex individuals. As shown for European white oaks, first-generation hybrids will also more likely mate with the more abundant species, leading to potential asymmetric introgression ([22]). Asymmetric introgression based on the abundance of the parental species has been described in other Mediterranean ([27]) and non-Mediterranean tree species ([1], [6], [10]).
Conclusions
These results support current bidirectional introgression with Q. suber and Q. ilex. Analysis of the progenies of four open pollinated hybrid trees from a natural mixed population reveals that each mother tree is predominantly fertilized by one of the two parental species, usually the most abundant in its vicinity. The frequency of backcrossing could at least be partially driven by the availability of pollen when female flowers are receptive. However, other post-pollination processes, such as genetic incompatibilities cannot be excluded. To clarify this point, controlled pollinations and monitoring of pollen tube development and abortion of embryos should be performed. Additionally, further investigations should take advantage of modern high-throughput sequencing methodologies that will allow the examination of larger proportions of the species’ genomes. Moreover, using these methodologies, it may be possible to identify the genome portions that determine hybridizing capability, and/or reproductive isolation in the Q. ilex - Q. suber complex.
Acknowledgements
AS and ULH conceived the idea, designed the experiments, collected the plant material and drafted the manuscript; HS and ULH genotyped the seedlings and adult trees; ULH, HS and AS performed the admixture and statistical analyses. All the authors have contributed to the final manuscript.
This work has been funded by the project AGL2015-67495-C2-2-R (Spanish Ministry of Economy and Competitiveness).
The authors thank the two anonymous reviewers for their helpful comments and suggestions.
References
Gscholar
Gscholar
Gscholar
CrossRef | Gscholar
Gscholar
Gscholar
CrossRef | Gscholar
Authors’ Info
Authors’ Affiliation
Héctor Sánchez
Alvaro Soto
GI Genética, Fisiología e Historia Forestal, Dpto. Sistemas y Recursos Naturales, ETSI Montes, Forestal y del Medio Natural, Universidad Politécnica de Madrid, Madrid (Spain)
Corresponding author
Paper Info
Citation
López De Heredia U, Sánchez H, Soto A (2018). Molecular evidence of bidirectional introgression between Quercus suber and Quercus ilex. iForest 11: 338-343. - doi: 10.3832/ifor2570-011
Academic Editor
Piermaria Corona
Paper history
Received: Jul 28, 2017
Accepted: Feb 20, 2018
First online: Apr 18, 2018
Publication Date: Apr 30, 2018
Publication Time: 1.90 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2018
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 46827
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 39676
Abstract Page Views: 2915
PDF Downloads: 3253
Citation/Reference Downloads: 9
XML Downloads: 974
Web Metrics
Days since publication: 2872
Overall contacts: 46827
Avg. contacts per week: 114.13
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2018): 11
Average cites per year: 1.38
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Fine-scale spatial genetic structure in a multi-oak-species (Quercus spp.) forest
vol. 8, pp. 324-332 (online: 05 September 2014)
Research Articles
SimHyb: a simulation software for the study of the evolution of hybridizing populations. Application to Quercus ilex and Q. suber suggests hybridization could be underestimated
vol. 11, pp. 99-103 (online: 31 January 2018)
Research Articles
Leaf morphology of progenies in Q. suber, Q. ilex, and their hybrids using multivariate and geometric morphometric analysis
vol. 11, pp. 90-98 (online: 31 January 2018)
Research Articles
Chloroplast microsatellites as a tool for phylogeographic studies: the case of white oaks in Poland
vol. 8, pp. 765-771 (online: 19 July 2015)
Research Articles
Clonal structure and high genetic diversity at peripheral populations of Sorbus torminalis (L.) Crantz.
vol. 9, pp. 892-900 (online: 29 May 2016)
Research Articles
Seedling emergence capacity and morphological traits are under strong genetic control in the resin tree Pinus oocarpa
vol. 17, pp. 245-251 (online: 16 August 2024)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Patterns of genetic variation in bud flushing of Abies alba populations
vol. 11, pp. 284-290 (online: 13 April 2018)
Research Articles
Comparison of range-wide chloroplast microsatellite and needle trait variation patterns in Pinus mugo Turra (dwarf mountain pine)
vol. 10, pp. 250-258 (online: 11 February 2017)
Research Articles
Genetic diversity of core vs. peripheral Norway spruce native populations at a local scale in Slovenia
vol. 11, pp. 104-110 (online: 31 January 2018)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword