Predicting total and component biomass of Chinese fir using a forecast combination method
iForest - Biogeosciences and Forestry, Volume 10, Issue 4, Pages 687-691 (2017)
doi: https://doi.org/10.3832/ifor2243-010
Published: Jul 17, 2017 - Copyright © 2017 SISEF
Research Articles
Abstract
Accurate estimates of tree biomass are critical for forest managers to assess carbon stock. Biomass of Chinese fir (Cunninghamia lanceolata [Lamb.] Hook.) in southern China was assessed by three alternative methods. In the Separate model approach, total and component tree biomass was directly predicted from a regression equation as a function of tree diameter and height. In the Additive model approach, total biomass was predicted as the sum of predictions from all component biomass equations. The Forecast Combination method involved combining predictions from the total biomass equation with the sum of predictions from component biomass equations. Results indicated that the Separate model method outperformed the Additive model method in predicting total and component biomass. The drawback of the Separate model method is that the total is not equal to the sum of its components. The Forecast Combination method provided the overall best prediction for total and component biomass, and still ensured additivity of component biomass predictions.
Keywords
Additivity, Biomass Predictions, Cunninghamia lanceolata, Even-aged Plantations, Tree Allometry
Introduction
Forest biomass comprises the arboreal fraction of all existing plant mass in the forest, including stems, branches, leaves, and roots of forest trees ([19]). Due to its important role as carbon pool in forest ecosystems ([6]) and the laborious and costly process of measuring trees in the forest, there is a great demand to accurately predict tree biomass. A number of approaches for tree biomass prediction have been put forward. A widely used approach is the direct prediction of biomass using allometric relationships from tree measurements, such as diameter at breast height, total tree height, crown radius, and wood density ([4], [23], [7], [11]).
There are two methods for predicting total tree biomass with allometric equations: tree-level and component-level. The tree-level method involves a regression to predict total tree biomass. In the component-level method, prediction of total tree biomass is the sum of predictions of all tree components (leaves, branches, stem, and roots), obtained from separate regressions. There are strengths and weaknesses for each method. The tree-level biomass model predicts total tree biomass directly, but lacks detailed information on biomass of stems, branches, leaves, and roots. On the other hand, the component-level method provides more detailed information, but total tree biomass obtained by summing component predictions could often suffer from accumulation of errors and subsequently poor accuracy and precision. Moreover, in the component-level method, the sum of the biomass components can generate inconsistent results, as compared to predictions from the total biomass model ([16], [19]). To eliminate this inconsistency, several model estimation methods have been suggested to enforce additivity on a system of biomass equations ([8], [17], [20], [15], [3]).
Forecast combination, introduced by Bates & Granger ([1]), is a method to improve forecast accuracy ([14]). This method combines information generated from different models and distributes errors from these models, thus ensuring consistency of outputs from different models. Zhang et al. ([24]) used this method to combine tree-level and stand-level predictions of stand basal area.
The objective of this study was to evaluate current methods of predicting total and component biomass against the forecast combination method.
Materials and methods
Study sites
The plantations studied were at Weimin farm (Shaowu city, Fujian province) and Nianzhu farm (Fenyi city, Jiangxi province) in southern China (Fig. 1). Both sites belong to the subtropical monsoon climate region. In Weimin farm, mean annual precipitation is 1768 mm, mean annual temperature is 17.7 °C, and monthly mean temperature ranges from 6.8 °C in January to 28 °C in July. In Nianzhu farm, mean annual temperature and precipitation are 17.2 °C and 1656 mm, respectively.
Weimin farm consisted of stands of 7-, 16- and 28-year-old Chinese firs (Cunninghamia lanceolata [Lamb.] Hook.). Nianzhu farm consisted of stands of 28-year-old Chinese firs. One or two trees in each diameter class (2-cm) in each plot (0.06 ha) were randomly destructively sampled, totaling 39 sample trees in Weimin farm and 24 trees in Nianzhu farm (Tab. 1).
Tab. 1 - Mean and standard deviation (SD) for tree variables and component biomass of Chinese fir by location and age. Values in parentheses are ranges.
| Location | Age | n | Stats | D (cm) | H (m) | Stem (kg) |
Branch (kg) |
Leaf (kg) |
Root (kg) |
|---|---|---|---|---|---|---|---|---|---|
| Weimin | 7 | 9 | Mean | 10.97 (5.7-16.3) | 7.28 (4.9-9.3) | 14.67 | 3.60 | 5.25 | 6.97 |
| SD | 3.84 | 1.61 | 9.08 | 2.41 | 4.25 | 5.56 | |||
| 16 | 14 | Mean | 14.19 (5.6-22.5) | 11.48 (5.9-14.8) | 34.26 | 2.63 | 3.45 | 11.38 | |
| SD | 5.28 | 2.70 | 22.57 | 2.50 | 3.18 | 8.90 | |||
| 28 | 16 | Mean | 16.86 (8.7-27.8) | 17.07 (10.3-22.7) | 71.11 | 5.72 | 4.22 | 18.41 | |
| SD | 5.82 | 3.30 | 51.04 | 7.16 | 3.68 | 16.03 | |||
| Nianzhu | 28 | 24 | Mean | 18.70 (7.5-30.2) | 16.89 (10.2-23.2) | 86.67 | 6.82 | 4.65 | 21.26 |
| SD | 7.13 | 3.57 | 64.90 | 6.97 | 4.53 | 18.18 |
Tree diameter at breast height (D) and height (H) was measured after the tree was felled. The fresh and dry weights of stem, branch, leaf and root were determined separately. From each stem, we sampled a 5-cm thick stem disc cut from the base of each 1-m stem segment, three subsamples of the branches and leaves from the upper, middle, and lower crown (1/3 of crown length, approximately 500-1000 g each), and samples (500-1000 g) from the stump, large structural roots (more than 10 mm), small roots (2-10 mm), and fine roots (less then 2 mm), obtained after excavation of the whole root system to the extent of the crown projection area. All samples were fresh weighted in the field, then transported to the laboratory where they were oven-dried to a constant weight at 105 °C and dry weighted. Dry weight was computed for each tree component by extrapolating the ratio of dry weight to fresh weight from subsamples.
Allometric equations
Standard allometric equations predict tree biomass as a power function of D ([11]). Other variables, such as total tree height, have also been proven to be important predictors ([4], [13]). The following two widely used equations ([12], [10], [23], [27], [21]) were considered in this study (eqn. 1, eqn. 2):
where Mi is the biomass (kg) for the i-th tree, a and b are the regression parameters to be estimated, Di is the tree diameter (cm), Hi is the total height (m), and εi is the random error.
The separate model method involves employing separate regression models to predict total tree biomass and its components: branch, leaf, root, and stem. Based on a preliminary analysis, we found that the eqn. 1 performed better than eqn. 2 on modeling branch, leaf and root biomass. However, in terms of the stem and total biomass, the eqn. 2 was better than eqn. 1. Thus, the following equation forms were selected (eqn. 3 to eqn. 7):
where all variables are as defined earlier, with added subscripts to denote component types.
The models were separate in the sense that prediction of total tree biomass from eqn. 7 did not equal the sum of predictions for components from eqn. 3 to 6. In other words, the equations were not constrained to be additive. Parameters of eqn. 3 to 7 were simultaneously obtained by using Seemingly Unrelated Regression (SUR). SAS procedure MODEL ([18]) was used for this purpose.
The Additive model approach was based on the procedure developed by Parresol ([15]). The following system of equations was used to predict total tree biomass and its components (eqn. 8 to eqn. 12):
where the symbol ^ on top of a variable name denotes the predicted value for that variable.
Eqn. 8 to 11 have the same forms as eqn. 3 to 6, respectively. Prediction of total tree biomass (eqn. 12) was obtained by summing predictions of component biomass. Again, SUR was used to estimate parameters of this system of equations.
Zhang et al. ([24], [25]) applied the forecast combination method to combine different types of models for predicting stand basal area and stand survival. A similar approach was applied in this study to predict total tree biomass by combining: (a) direct prediction from the regression model (eqn. 7); and (b) indirect prediction by summing predictions from component models (eqn. 13):
where {hat} MFi is the prediction of total tree biomass from forecast combination, w1 and w2, are weights, with w1 + w2 = 1, {hat}MTi is the direct prediction of total tree biomass from eqn. 7, and {hat} MTSi is the indirect prediction of total tree biomass by summing predictions from component biomass equations. Depending on the component biomass equations, two methods of Forecast Combination were considered in this study: (i) FC1 that used the sum of predictions from the Separate model (eqn. 3-6); and (ii) FC2 that used the sum of predictions from the Additive model (eqn. 12). The weight coefficients of the combined tree biomass model could be obtained by the optimal weight method ([24] - eqn. 14):
where εki is the prediction error for the tree i using the method k, with k = 1, 2, and i = 1, 2, …, m, being m the total number of trees; T is the transport matrix.
The component predictions were then adjusted to add up to the combined estimator for total tree biomass. This was accomplished by multiplying the component predictions by λ, the adjusting coefficient, which is calculated as (eqn. 19):
In this study, the two-fold leave-one-out cross validation scheme was used for model validation. First, the models were fitted using data from the Weimin farm, and then validated using data from the Nianzhu farm. Second, we treated Nianzhu data as the fit data and Weimin data the validation data. Evaluation statistics were computed based on observations pooled from the two validation data sets. The evaluation statistics of mean difference (MD), mean absolute difference (MAD), and R2 ([24]) were used to validate the models. Models with lower MD and MAD values indicate a better fit to the data.
Results and discussion
The stem biomass ranged from 14.67 to 86.67 kg, branches from 2.63 to 6.82 kg, leaves from 3.45 to 5.25 kg, and roots from 6.77 to 21.23 kg (Tab. 1). Stem biomass accounted for more than 50% of the total tree biomass, except the stand of age 7, which could explain the same equation (eqn. 2) of stem and total tree biomass in the preliminary analysis. The parameter estimates and their standard deviation errors were slightly different between the Separate model and Additive models in the Weimin and Nianzhu farms (Tab. 2), being their values more consistent across methods than across farms. For total tree biomass prediction, the MD value obtained using the FC1 method (the Forecast Combination method that used separate models) was 83.46% smaller than that obtained using the additive model and 41.04% smaller than that of the FC2 method (Forecast Combination method that used additive model). Moreover, the FC1 method produced the best R2 values, while the Separate model method yielded the lowest MAD value (Tab. 3). Regarding the prediction of component biomass, the FC1 method had the best MAD and R2 values for all components (except leaf biomass) and the best MD values for two of the four components (Tab. 4).
Tab. 2 - Parameter estimates and standard errors (SE) of the biomass model using Separate and Additive models in the Weimin and Nianzhu farms.
| Parameter | Weimin | Nianzhu | ||
|---|---|---|---|---|
| Estimates | SE | Estimates | SE | |
| a 3B | 8.2e-5 | 6.4e-5 | 8.84e-4 | 0.0013 |
| b 3B | 3.7571 | 0.2425 | 2.9377 | 0.4520 |
| a 4L | 0.0276 | 0.0215 | 0.0009 | 0.0013 |
| b 4L | 1.7984 | 0.2574 | 2.7953 | 0.4455 |
| a 5R | 0.0110 | 0.0047 | 0.0200 | 0.0103 |
| b 5R | 2.5401 | 0.1383 | 2.3165 | 0.2503 |
| a 6S | 0.0247 | 0.0053 | 0.0397 | 0.0131 |
| b 6S | 0.9141 | 0.0236 | 0.8647 | 0.0346 |
| a 7T | 0.0337 | 0.0066 | 0.0435 | 0.0156 |
| b 7T | 0.9211 | 0.0215 | 0.8897 | 0.0376 |
| a 8B | 0.0001 | 8.6e-5 | 0.0015 | 0.0021 |
| b 8B | 3.7089 | 0.2966 | 2.7895 | 0.4583 |
| a 9L | 0.0432 | 0.0396 | 0.0010 | 0.0015 |
| b 9L | 1.6763 | 0.3055 | 2.7802 | 0.4539 |
| a 10R | 0.0089 | 0.0041 | 0.0114 | 0.0058 |
| b 10R | 2.6206 | 0.1480 | 2.4950 | 0.2641 |
| a 11S | 0.0224 | 0.0049 | 0.0369 | 0.0121 |
| b 11S | 0.9254 | 0.0239 | 0.8726 | 0.0344 |
Tab. 3 - Evaluation statistics for total tree biomass prediction by method. (§): denotes the best method based on each fitting statistic (MD, MAD, R2).
| Method | MD | MAD | R2 |
|---|---|---|---|
| Separate model | 0.5761 | 8.0296(§) | 0.9772 |
| Additive model | -0.7199 | 8.6210 | 0.9726 |
| Combined using Separate model | 0.1191(§) | 8.0469 | 0.9780(§) |
| Combined using Additive model | 0.2020 | 8.0802 | 0.9776 |
Tab. 4 - Evaluation statistics for component biomass prediction by method. (§): denotes the best method based on each fitting statistic (MD, MAD, R2).
| Component | Method | MD | MAD | R2 |
|---|---|---|---|---|
| Branch | Separate model | -0.1511 | 1.9476 | 0.6601 |
| Additive model | -0.487 | 2.0743 | 0.6371 | |
| Combined using Separate model | -0.0853(§) | 1.9307(§) | 0.6786(§) | |
| Combined using Additive model | -0.3997 | 2.0213 | 0.6755 | |
| Leaf | Separate model | 0.7034(§) | 2.0181(§) | 0.4952(§) |
| Additive model | 0.4883 | 2.0838 | 0.4424 | |
| Combined using Separate model | 0.7343 | 2.0768 | 0.4332 | |
| Combined using Additive model | 0.5259 | 2.1002 | 0.4311 | |
| Root | Separate model | -0.2353 | 3.2619 | 0.8807 |
| Additive model | -0.2625 | 3.3982 | 0.8686 | |
| Combined using Separate model | -0.0046(§) | 3.2564(§) | 0.8905(§) | |
| Combined using Additive model | -0.0466 | 3.2853 | 0.8872 | |
| Stem | Separate model | -0.298 | 4.7543 | 0.9832 |
| Additive model | -0.4586 | 4.9985 | 0.9831 | |
| Combined using Separate model | 0.1986 | 4.2931(§) | 0.9865(§) | |
| Combined using Additive model | 0.1224(§) | 4.5509 | 0.9851 |
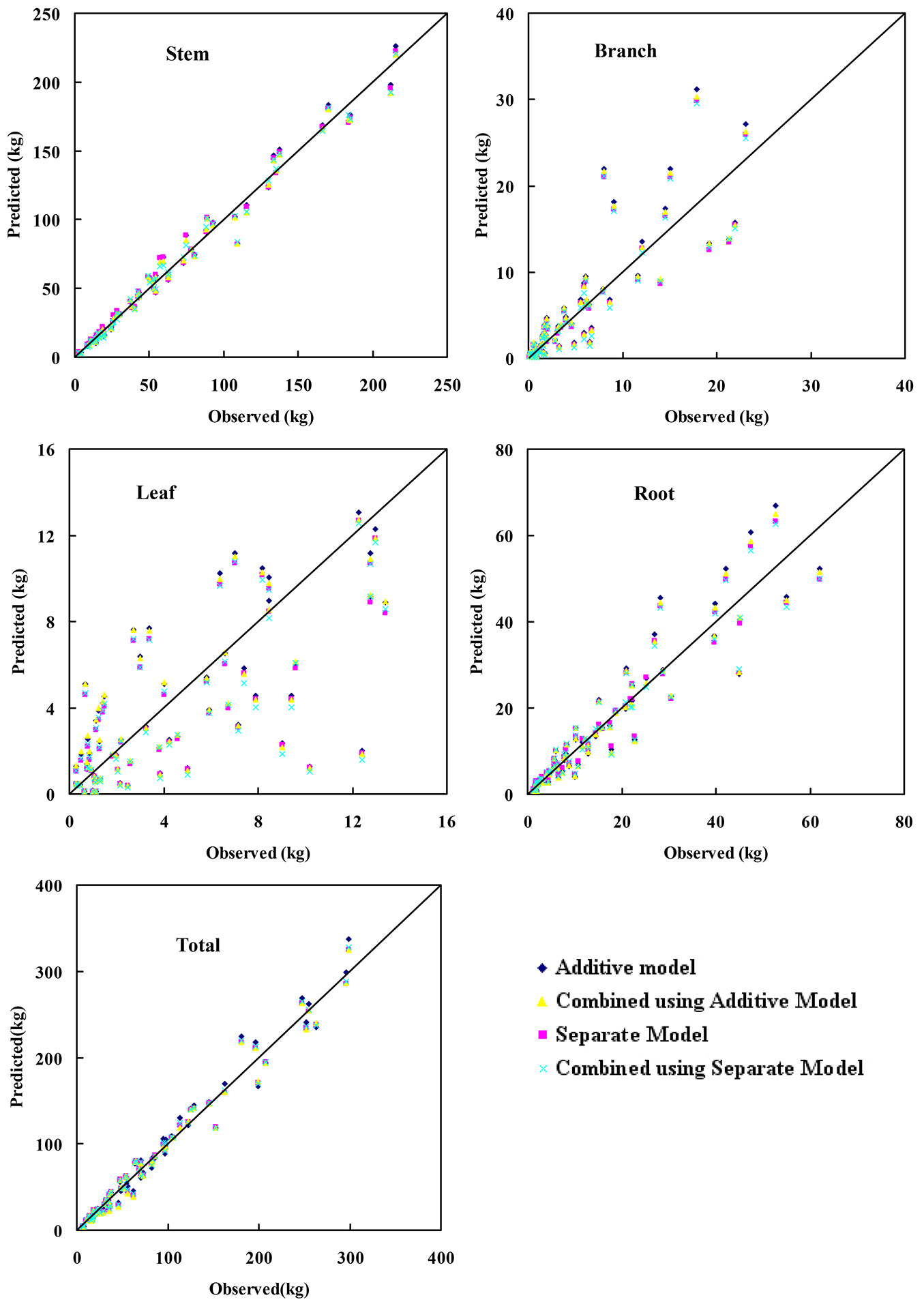
Fig. 2 displays the predicted vs. observed biomass of total and component biomass. Most of biomass predictions were distributed near the straight line (y=x) using any of the four methods used (Fig. 2). It can be noticed that the predictions of leaf biomass showed the lowest accuracy and precision among all the component biomass.
Additive vs. separate model
Parameters of the component regression models in the Additive model approach were subjected to the constraint that the sum of component predictions from the resulting models would be equal to the total biomass ([9], [3], [5]). On the other hand, the Separate model method did not involve any constraints, thus its parameters resulted 19.97% and 6.86% lower for MD, MAD, respectively and 0.47% larger for R2 than those of the Additive model method used for modeling total tree biomass (Tab. 3). Furthermore, the separate model method produced the best MD, MAD and R2 values for all components. The sole exception using the latter method was leaf biomass, which showed a lower MD value (Tab. 4). However, due to lack of additivity constraint, the sum of predictions from different biomass components was not equal to the prediction of total tree biomass in the Separate model approach ([19]). Therefore, the advantage of a compatible, additive system for total and component biomass in the Additive model method was also accompanied by a decrease in the accuracy and precision of predictions.
Forecast combination
This approach involves a combined estimator, which is the weighted average of the predictions from the total biomass equation and the sum of predictions of component biomass from either (a) the separate model, or (b) the additive model. Yue et al. ([22]) used the variance-covariance method to calculate weight coefficients for the combined model. Zhang et al. ([26]) reported that the optimal weight method (i.e., the ordinary least squares estimate of the weights) performed better than the variance-covariance method. Here, the optimal weight method was used to calculate weight coefficients of combined model of tree biomass. Compared to the method using the additive model (FC2), the Forecast Combination method that used the separate model (FC1) gave better values of MD, MAD, and R2 for total and component biomass prediction (Tab. 3, Tab. 4). The only exceptions were MD values for leaf and stem biomass. The sum of component predictions from the Separate model was not subject to any constraint, and therefore could be different from the prediction from the total biomass equation. The combined predictions of FC1 method benefit from non-constraint of the Separate model, thus increasing the accuracy and precision as compared to the FC2 estimation.
Forecast combination vs. additive model
Tree biomass additivity has long been recognized as a desirable property of biomass estimation. Several studies have successfully solved the logical inconsistency between the components and total tree predictions ([17]) by developing a system of additive biomass equations estimated by the SUR method, thus providing a statistically correlated system of equations with restrictions ([15], [2]). In this study, a forecast combination with adjusted coefficient (a consistent value) was applied, ensuring the additivity of the component biomass. It is worth noting that the adjusted coefficient may vary across the biomass components, because the ratio between each component depends on site quality, stand density, age or tree size.
The FC1 method outperformed the Additive model method in predicting the total tree biomass, based on all three evaluation statistics. The FC1 method also produced better values of MD, MAD, and R2 for predicting branch, root, and stem biomass. For leaf biomass, the FC1 method resulted in better MAD, but worse MD and R2 values, as compared to the Additive model method (Tab. 3, Tab. 4).
Forecast combination vs. separate model
In the Separate model method, the prediction of total tree biomass for each tree is directly derived from the total biomass regression equation. The FC1 method combined the information from this prediction and the sum of predictions from regression equations for each component biomass. The result is an improvement of two (MD and R2) out of three fitting statistics for total tree biomass predictions obtained using the FC1 method, as compared with the Separate model method (Tab. 3). Indeed, the MD value derived from FC1 method was 79.33% smaller than that obtained using the Separate model method, while the R2 value from FC1 method was slightly larger than that of the Separate model method.
For the component biomass, predictions from the Separate model method were unadjusted predictions from the regression models. These predictions were then adjusted such that the resulting sum matched the combined estimator for total biomass in the FC1 method.
Based on all three evaluation statistics, the FC1 method showed better performances in predicting branch, root, and stem biomass as compared to the Separate model, whereas the latter method yielded more accurate predictions of leaf biomass. The opposite trend observed in leaf biomass predictions might be due to low R2 values of separate leaf biomass model (Tab. 4). In the forecast combination method, the implicit assumption was that the relationship between observed and predicted values from different models was stable. If this relationship remains relatively unchanged from the sample data to the population, then the combined value should provide better predictions than those by any model alone ([26]). However, in this study the relationship in leaf biomass was not stable, as inferred from R2 values ranging from 0.43 to 0.49 (Tab. 4).
Conclusions
The Forecast Combination method takes advantages of information from tree-level and component-level models, by providing an estimator that combines predictions from these models. To ensure additivity, component predictions from the Separate model were adjusted to match the combined estimator for total tree biomass. This approach was superior to the Additive model method in predicting total tree biomass and all of its components, except for leaf biomass.
Acknowledgments
The authors gratefully acknowledge the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (CAFYBB2017ZX001-2), the National Natural Science Foundation of China (No. 31670634), and the Scientific and Technological Task in China (No. 2016YFD0600302-1).
References
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Congwei Xiang
Aiguo Duan
Jianguo Zhang
State Key Laboratory of Tree Genetics and Breeding, Key Laboratory of Tree Breeding and Cultivation of the State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing 100091 (P. R. China)
Congwei Xiang
Aiguo Duan
Jianguo Zhang
Collaborative Innovation Center of Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing (P.R. China)
School of Renewable Natural Resources, Louisiana State University, Agricultural Center, Baton Rouge, LA 70803 (USA)
Corresponding author
Paper Info
Citation
Zhang X, Cao QV, Xiang C, Duan A, Zhang J (2017). Predicting total and component biomass of Chinese fir using a forecast combination method. iForest 10: 687-691. - doi: 10.3832/ifor2243-010
Academic Editor
Matteo Garbarino
Paper history
Received: Oct 08, 2016
Accepted: May 16, 2017
First online: Jul 17, 2017
Publication Date: Aug 31, 2017
Publication Time: 2.07 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2017
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 48580
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 40615
Abstract Page Views: 2899
PDF Downloads: 4074
Citation/Reference Downloads: 28
XML Downloads: 964
Web Metrics
Days since publication: 3141
Overall contacts: 48580
Avg. contacts per week: 108.26
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2017): 10
Average cites per year: 1.11
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Relationship between volatile organic compounds released and growth of Cunninghamia lanceolata roots under low-phosphorus conditions
vol. 11, pp. 713-720 (online: 06 November 2018)
Research Articles
Mechanical and physical properties of Cunninghamia lanceolata wood decayed by brown rot
vol. 12, pp. 317-322 (online: 06 June 2019)
Research Articles
Integration of tree allometry rules to treetops detection and tree crowns delineation using airborne lidar data
vol. 10, pp. 459-467 (online: 04 April 2017)
Research Articles
Tree biomass models for the entire production cycle of Quercus suber
vol. 18, pp. 38-44 (online: 28 February 2025)
Research Articles
Soil CO2 efflux in uneven-aged and even-aged Norway spruce stands in southern Finland
vol. 11, pp. 705-712 (online: 06 November 2018)
Research Articles
Aboveground tree biomass of Araucaria araucana in southern Chile: measurements and multi-objective optimization of biomass models
vol. 14, pp. 61-70 (online: 09 February 2021)
Research Articles
Deriving tree growth models from stand models based on the self-thinning rule of Chinese fir plantations
vol. 15, pp. 1-7 (online: 13 January 2022)
Research Articles
High resolution biomass mapping in tropical forests with LiDAR-derived Digital Models: Poás Volcano National Park (Costa Rica)
vol. 10, pp. 259-266 (online: 23 February 2017)
Technical Reports
Biomass equations for European beech growing on dry sites
vol. 9, pp. 751-757 (online: 17 June 2016)
Research Articles
On the geometry and allometry of big-buttressed trees - a challenge for forest monitoring: new insights from 3D-modeling with terrestrial laser scanning
vol. 8, pp. 574-581 (online: 02 March 2015)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword