Comparison of parametric and nonparametric methods for modeling height-diameter relationships
iForest - Biogeosciences and Forestry, Volume 10, Issue 1, Pages 1-8 (2016)
doi: https://doi.org/10.3832/ifor1928-009
Published: Oct 19, 2016 - Copyright © 2016 SISEF
Research Articles
Abstract
This paper focuses on the problem of regionalization of the height-diameter model at the stand level. To this purpose, we selected two different modeling techniques. As a parametric method, we chose a linear mixed effects model (LME) with calibrated conditional prediction, whose calibration was carried out on randomly selected trees either close to mean diameter or within three diameter intervals throughout the diameter range. As a nonparametric method, the technique of classification and regression trees (CART) was chosen. These two methods were also compared with the local model created by ordinary least squares regression. The results show that LME with calibrated conditional prediction based on measurements of height at three diameter intervals provided results very close to the local model, especially when six to nine trees are measured. We recommend this technique for the regionalization of the global model. The CART method provided worse results than LME, with the exception of parameters of the residual distribution. Nevertheless, the latter approach is very user-friendly, as the regression tree creation and especially its interpretation are relatively simple, and could be recommended when larger deviations are allowed.
Keywords
Calibration, Classification and Regression Trees, Hierarchical Structure, Linear Mixed Effects Model, Spatial Heterogeneity
Introduction
Modeling the relationship between the tree height and its diameter at breast height is one of the most common and oldest paradigms in forestry, especially in forest inventory ([37]). Various linear and nonlinear models have been used to study such relationship ([48]). Estimates are usually obtained by the ordinary least squares (OLS) method, though the estimation of model parameters is often hampered by the failure in satisfying the assumptions of the method. Indeed, the independence between observations is very often violated due to the spatial or temporal dependencies between data, leading to biased estimates of standard errors of parameters and their confidence intervals, as well as to misleading results of significance tests ([17]). One of the main causes of the dependence of data in forestry models is their hierarchical structure. Data collected from the same inventory units are more similar to each other than data from different inventory units ([17]). To face this problem, we must consider that spatial variability is an important feature of the ecosystem, rather than a mere statistical nuisance ([22]), and its accurate assessment allows to better understand the complexity and structure of an ecosystem ([49]). For this reason, models must aim to explain the spatial variability of data as much as possible. In previous studies, researchers often disregarded the problem of spatial and temporal non-stationarity, using global or local models instead. The real spatial structure is commonly neglected by global models, as they assume temporal and spatial homogeneity of the data, and this results in a high portion of unexplained variation. On the contrary, local models cannot be generalized to populations (e.g., large areas) - as fitting many individual models requires a large number of measurements - and are unsuitable to model the variability among different plots. Several authors reported different ways to overcome these drawbacks ([8]). Ferguson & Leech ([16]) used two-stage models, Huang et al. ([20]) applied models for each geographical or ecologically defined area, while Zhang et al. ([47]) used various geostatistical methods. Among nonparametric methods used on spatially correlated data, kriging has become very popular in the last decades; several examples are reported in Kangas & Haara ([21]).
The use of models with mixed effects is a successful approach to overcome the problem of spatial variability in the data. Mixed models include a fixed component, which covers the entire analyzed dataset (e.g., population), and a random component that involves various levels of hierarchy in the dataset ([8]). Mixed effects model allows the simultaneous estimation of parameters of the fixed and random components, thus combining the “average” characteristics of the analyzed population (through fixed parameters of the global model) with the spatial or temporal variability of data (through the random component parameters - [2]). The fixed component of the model therefore explains the influence of different variables as in the classical OLS regression ([46]). In contrast, the random component takes into account the heterogeneity and randomness in the data caused by known and unknown factors ([44]). Mixed models thus provide a better parameter estimates than OLS regressions ([7]), providing a flexible tool to analyze a given area.
The main advantage of using a mixed model is the possibility to calibrate an already existing model ([26]) by estimating its parameters on data not used in the construction of the original model, as in the case of a new experimental plot. Two calibration methods are available to this purpose ([38]):
- Random parameters have a value of 0. There are no measured values of the dependent variable, so the calibration is done with only fixed effects (often referred to as “fixed effects marginal prediction”), which is related to the entire population.
- Random parameters are estimated separately for each sampling unit. It includes at least one (or better several) measured values of the dependent variable. It is a calibration with fixed and random effects (also referred to as “calibrated conditional prediction”), which applies to individual areas.
These calibration techniques have a wide range of applications, though so far they have been most frequently used in modeling growth ([28]), increment ([8], [7], [1]) and height-diameter functions ([42], [2], [37], [9], [24]).
When trees in the experimental area are not enough for applying the calibrated conditional prediction, LME with fixed effects marginal predictions has to be used for tree height modeling, instead of the LME model with calibrated conditional predictions. In such cases, a suitable alternative could be the method of classification and regression trees (CART), which is a nonparametric technique that effectively use local data ([40]), does not require a normal distribution of residuals ([15]), and is robust to outliers ([11]). According to Rejwan et al. ([35]), this method is suitable for modeling with hierarchical data. This approach is based on the splitting of binary data into homogeneous groups (with respect to the dependent variable) by using independent variables ([12]). Using qualitative dependent variables, this method produces a classification tree, while a regression tree is obtained using quantitative dependent variables ([29]). The main advantage of this approach is that no additional height measurements are required for modeling tree height at the local level when a CART model based on other data sets is available for the area of interest. In this case, only the stand variables required for the assembled classification or the regression tree have to be identified.
Although far less used in forestry than the LME, the CART method has been successfully applied for modeling tree mortality ([12]) and for the regionalization of global models of the volume of tally trees ([33]) or of floodplain forest occurrences ([5]). Therefore, an accurate evaluation its advantages and disadvantages in comparison with the widely used LME method is required.
The aim of this paper is to compare the use of the LME (parametric) and the CART (nonparametric) models to regionalize the global model of height-diameter relationship.
Material and methods
Study area
The study site is located in the Training Forest Enterprise Masaryk Forest Krtiny (lat. 49° 17′ 43″ N, long. 16°45′ 01″ E), southern Moravia (Czech Republic), north of Brno. Forty-six circular sample plots were selected in 23 different Norway spruce (Picea abies Karst.) stands. The sample plot area was 250 m2 (stands younger than 40 years), 700 m2 (stands aged between 41-80 years) and 1200 m2 (stands older than 80 years). For each tree in the sample plots, its height and diameter at breast height were measured, and the basal area of each tree was calculated. Volume of each tree was calculated using volume equations by Petráš & Pajtík ([32]). We measured a total of 1.590 trees. For each of the 23 forest stands considered, the mean diameter, mean height, stand basal area, mean tree volume, number of trees per ha, site index (mean height in 100 years) and the stand age were obtained by averaging the sampling plot values. Tab. 1 summarizes the basic characteristics of tree and stand variables over the study area.
Tab. 1 - Basic characteristics of tree and stand variables. (DBH): diameter at breast height of a tree; (H): height; (BA): basal area; (V): volume; (DBHm): mean diameter; (Hm): mean height; (BAs): stand basal area; (Vm): mean tree volume; (N): number of trees; (SI): site index; (A): stand age; (95% CI): 95% confidence interval of mean value; (min): minimal value; (Max): maximal value; (STD): standard deviation.
| Level | Variable | Mean ± 95% CI | Min | Max | STD |
|---|---|---|---|---|---|
| Tree | DBH (cm) | 32.2 ± 0.5 | 9.0 | 75.0 | 10.9 |
| H (m) | 28.4 ± 0.3 | 10.0 | 41.5 | 6.4 | |
| BA (cm2) | 911.0 ± 29.7 | 63.6 | 4417 | 603 | |
| V (m3) | 1.17 ± 0.04 | 0.01 | 6.55 | 0.91 | |
| Stand | DBHm (cm) | 33.1 ± 0.4 | 18.6 | 47.4 | 8.3 |
| Hm (m) | 28.8 ± 2.8 | 14.6 | 36.5 | 6.4 | |
| BAs (m2 ha-1) | 35.7 ± 0.3 | 22.2 | 46.3 | 6.34 | |
| Vm (m3) | 1.17 ± 0.03 | 0.16 | 2.56 | 0.66 | |
| N (n ha-1) | 498.0 ± 13 | 221 | 1240 | 272 | |
| SI (m) | 33.6 ± 0.1 | 32 | 38 | 1.7 | |
| A (years) | 81.6 ± 1.6 | 30 | 136 | 33.0 |
Statistical analysis
All the analyses were carried out using the software packages R ([34]) and STATISTICA® v. 12.0 (StatSoft Inc., Tulsa, OK, USA). The performances of the parametric LME and the nonparametric CART methods in modeling the height-diameter relationship were compared. We fitted the global model of the height-diameter function for the entire surveyed area and local models for each forest stand. To fit a model with mixed effects, an appropriate height-diameter (H-D) function has to be selected. We have evaluated five common H-D functions: (i) the Michailov function ([27] - eqn. 1):
(ii) the Petterson function ([31] - eqn. 2):
(iii) the Näslund function ([30] - eqn. 3):
(iv) the Levakovic function ([23] - eqn. 4):
(v) the Meyer function ([25] - eqn. 5):
where h is the fitted height of a tree, dbh is the diameter at breast height, and a, b, c are the parameters to be estimated.
To select the best function for the global model, the following criteria were considered: (i) mean of residuals; (ii) residual standard deviation; (iii) standard error of residuals; (iv) Akaike’s information criterion (AIC - [4]).
Linear mixed effects model
We built the actual mixed model as a two-level model, according to the methodology proposed by Weiss ([45]). The first-level model included only tree variables, which vary within the forest stand in relation to height and diameter. The second-level model also included stand variables. In terms of general matrix expression, the linear mixed model can be described as follows ([19] - eqn. 6):
where y is the dimensional vector of the n observations of the response variable taken from nv trees within nu plots, X is the design matrix of dimension n × m including the explanatory variables associated with fixed components, β is the m-dimensional vector of fixed components (estimated fixed parameters of the model), Z is the design matrix of dimension n × q for the random components, b is the q-dimensional vector of random components for the levels tree and plot, and ε is the n-dimensional vector of residual components.
Making the first-level model consists of three steps: (1) the global model; (2) the model with random effect of intercept only; and (3) the model with random effect of intercept and regression parameter. Through these steps, we tested the significance of the structure in the data, the importance of the predictor, and the random effect of the model. The significance of each model was tested using the likelihood ratio test and the AIC. After the first-level model was obtained, we fitted the second-level model by testing the contributions of the following stand variables: number of trees per ha, age of the forest stand, stand basal area, mean diameter, and the site index. In some cases, the logarithmic transformation was applied to stand variables because of the non-linear relationships observed between stand variables and the model parameter estimates.
The inclusion of the stand variables in the second-level model was done using the following procedure: the stand variable showing the highest correlation with the parameter estimates was included in the model; after the inclusion, the relation of the remaining stand variables on parameter estimation was tested again. We repeated the above procedure until the parameter estimates showed a statistically significant relationship (correlation coefficient) with some of the tested stand variables.
The mixed model obtained was calibrated using the calibrated conditional prediction in order to extend its predictability to the other plots. This step requires at least one dependent variable value to be measured in a given plot (in this case, the tree height) in order to calculate the random component of the parameter estimates. We can write the calibrated conditional prediction using the equation by Calama & Montero ([8] - eqn. 7):
where the meaning of the symbols is the same as in eqn. 6.
For the calculation of the random components of parameter estimation, we used the best linear unbiased predictor (BLUP) proposed by Robinson ([36]). The estimation of random components of the model parameters can be expressed as follows ([8] - eqn. 8):
where b is the vector of BLUP for the random components, D is the covariance matrix of the random effects, ZT is the design matrix for the random components, R is the estimated matrix for the residual variance, e is the vector whose dimension equals the number of observations and whose values are the residuals of the fixed effects marginal model.
The calibration process was carried out using two different procedures of tree selection. In the first procedure, we measured the height of 1 to 5 randomly selected trees that were in the range of ± 2 cm of the mean diameter. In the second case, three diameter (dmean) intervals of 4 cm were set as follows: (i) dmin to dmin + 4 cm; (ii) dmean - 2 cm to dmean + 2 cm; (iii) dmax - 4 cm to dmax. In this case, 1, 2 or 3 trees were randomly selected within each interval, obtaining 3, 6 or 9 trees, respectively, to be measured for height.
The calibration was performed on six forest stands that differed significantly in age and other stand characteristics. To assess the quality of the mixed model as compared with local models, we used the following criteria: (i) coefficient of determination (R2); (ii) root mean square error (RMSE); (iii) mean of residuals; (iv) residual standard deviation; (v) Akaike’s information criterion (AIC); (vi) mean deviation (Δi) of fitted values obtained by the LME model from the fitted values obtained by the OLS model, calculated as follows (eqn. 9):
where n is the sample size, yiLME and yiOLS are the fitted values obtained for the i-th tree (i = 1, 2, 3, …, n) by the LME and the OLS models, respectively.
Classification and regression trees
Because tree height is a continuous variable, the regression tree method was selected, whose results are homogeneous groups of individuals sharing the same predicted height (μh). Individual groups are explained using several different predictors, which are independent variables easy to be measured in the stand and/or to be computed from the measured variables. In this case, we used diameter at breast height, mean diameter of tree species, age of forest stand as continuous variables, as well as the site index as a categorical variable. We assessed the contribution of each variable to the regression tree grouping using the analysis of significance proposed by Breiman et al. ([6]). The degree of significance take values from 1 to 100; the larger the value, the greater the significance of given variable in the tree formation.
The regression tree model was tested on the same data used for the mixed model described above, but the original dataset was divided into a training (860 records) and a validation (730 records) subsets. The training subset was used to compile the regression tree model, while the validation subset to verify the accuracy of the model.
In the process of binary data splitting of the tree nodes, the algorithm tries to reduce the residual sum of squares. To prevent model overfitting and ensure a good estimate of μh, we set the minimum number of values in one node to 50, i.e., when one of the two groups obtained by splitting has fewer than 50 values, the process is terminated.
Ten-fold cross-validation pruning technique was chosen to reduce the number of branching of the regression trees achieved. After cross-validation, the regression tree models were tested on the validation subset. The quality of the resulting trees was determined using both subsets based on the following criteria: (i) coefficient of determination; (ii) mean of residuals; (iii) standard error of residuals; (iv) residual standard deviation.
Comparison of the results of the mixed model and regression tree
We compared the results of the mixed model and regression tree using data from six forest stands for which the mixed model calibration was carried out. In addition, the results of both methods were compared with those obtained with a local linear model of the height-diameter function fitted using the classical ordinary least squares (OLS) method. As comparison criteria, we have used the coefficient of determination (R2), mean of residuals (eÌi), the standard deviation of residuals (σei) and the mean deviation of fitted values obtained by the mixed model or the regression tree from the fitted values of the local model (Δi). For the comparison of the LME model with CART and OLS methods, we used only the mixed model with calibrated conditional prediction carried out with heights measured in three diameter intervals, as we have identified this type of calibration to show the best goodness-of-fit for the linear mixed effects model.
Results
Linear mixed effects model
Tab. 2 shows the results of the comparison of functions used in the global height-diameter models. Based on all the criteria chosen, the Petterson’s height-diameter function provided the best results. However, similar model performances were observed using all the functions tested, indicating their potential applicability in the prediction of tree height.
Tab. 2 - Comparison of global height-diameter models. (ei): mean of residuals; (σei): residual standard deviation; (SEei): standard error of residuals; (AIC): Akaike information criterion.
The resulting global linear model using the Petterson H-D function can be represented by the following formula (eqn. 10):
where h is the fitted height of the tree and dbh is its diameter at breast height. The whole model and its parameters were statistically significant (p<0.001), with a high coefficient of determination (R2 = 0.781).
All subsequent stages of mixed model (global model, the model with random effect of intercept only and model with random effect of intercept and regression parameter) were compared using the likelihood ratio test. Tab. 3 shows the results of all first-level sub-models. In all cases, we observed statistically significant differences among the different models. The final first-level mixed model therefore included the random part of the parameter estimation for both the intercept and the regression parameter.
Tab. 3 - Comparison of first-level models. (a, b): parameters of the model; (σ uai): standard deviation of the random intercept; (σ ubi): standard deviation of the random regression parameter; (εik): residual standard deviation; (AIC): Akaike’s information criterion; (p): p-value after the likelihood ratio test.
| Model | a | b | σ u ai | σ u bi | ε ik | AIC | p |
|---|---|---|---|---|---|---|---|
| Global | 0.2648 | 2.0848 | - | - | 0.0149 | -8861 | - |
| Random effect of a only | 0.2952 | 1.2624 | 0.0192 | - | 0.0076 | -10863 | < 0.001 |
| Random effect of a and b | 0.2944 | 1.2919 | 0.0194 | 0.1726 | 0.0001 | -10881 | < 0.001 |
When compiling the second-level model, we tested the contribution of individual stand variables. The obtained results suggest that the estimates of the parameter a are mainly dependent on the logarithm of the forest stand age (R = 0.944), while b estimates are dependent on the age of the forest stand (R = 0.457). Therefore, both forest stand age and its logarithmic transformation were included into the second-level model. Subsequently, the parameter estimates of the model including the stand age were tested for relationship with the remaining stand variables and no statistically significant correlations were found. Thus, the final mixed, second-level model included as predictors the stand age in parameter b and the logarithm of stand age in parameter a. We can express the final model with following equations (eqn. 11):
where (eqn. 12):
and (eqn. 13):
in which h is the fitted height of a tree, dbh is the diameter at breast height, T is the age of the forest stand, uai is the random part of the intercept, ubi is the random part of the regression parameter.
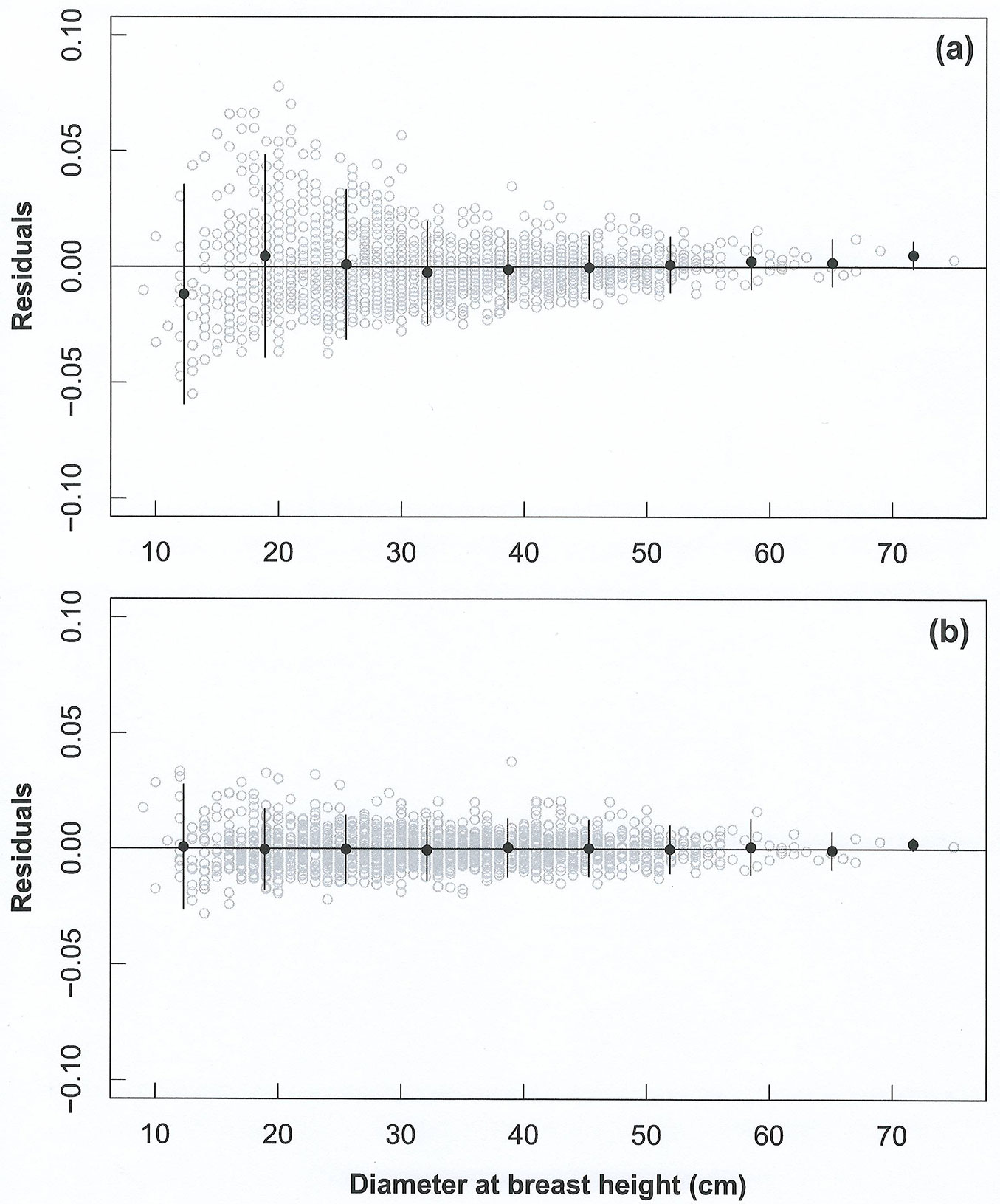
The second-level mixed-effects model showed a better fit on data as compared with the first-order model (likelihood ratio test: p < 0.001, AIC = -10927.7). This was confirmed by its lower dispersion of residuals, as depicted in Fig. 1.
Fig. 1 - Comparison of residuals (grey circles) of the global model (a) and the second-level mixed effects model (b) for the prediction of tree height. Black dots represent the mean of residuals in each diameter class, while the black lines represent its confidence intervals.
The calibrated conditional prediction of tree height based on a random sample of 1 to 5 trees within ± 2 cm around the mean diameter provided biologically inconsistent results. Indeed, an inverse relationship was found, thereby tree height was predicted to decrease when diameter increases. Such inconsistency could not be eliminated even by using a larger number of measurements (max 5). Due to the poor quality of this calibration, a low coefficient of determination was obtained, as well as a high average deviation of fitted values of the mixed model and fitted values of the local model.
The second calibration method (tree height measured in three diameter intervals - see above) yielded significantly better results. Tab. 4 summarizes the goodness-of-fit statistics obtained using the calibrated second-level mixed effects model on the whole data set (training + validation) for the six forest stands analyzed. The results clearly showed that increasing the numbers of height measurements (1, 2, or 3 trees per interval, i.e., 3, 6, and 9 trees), the accuracy of model predictions was largely improved. Indeed, a higher number of trees minimizes the impact of selecting dominant or suppressed trees on the position of the model curve. The results also show that the calibration using only three measured heights can provide confidence intervals of Δi higher than 1 m, which is obviously unacceptable from a practical point of view. On the contrary, Δi becomes significantly smaller when the model is run based on six or nine height measurements depending on the desired degree of accuracy, indicating the reliability of its predictions of tree height. We achieved the best results when the calibration was made using height measurements of nine trees. Subsequently, we used this model for comparison with the regression tree and the local model.
Tab. 4 - Goodness-of-fit of the calibrated second-level mixed effects model using 3 to 9 tree height measurements. (N): number of measured trees; (Δi): average deviation of fitted values obtained by the LME model and those obtained by the OLS model; (R2): coefficient of determination; (AIC): Akaike’s information criterion; (RMSE): root mean square error; (Ä i): mean of residuals; (σei): residual standard deviation; (95% CI): 95% confidence interval of the mean.
| Statistics | N | Forest stand no. | Mean (±95% CI) |
|||||
|---|---|---|---|---|---|---|---|---|
| 3 | 5 | 11 | 14 | 19 | 22 | |||
Δ i |
3 | 0.549 | 0.752 | 1.144 | 1.032 | 0.694 | 1.315 | 0.914 ± 0.310 |
| 6 | 0.542 | 0.339 | 0.665 | 0.764 | 0.590 | 1.115 | 0.669 ± 0.295 | |
| 9 | 0.400 | 0.239 | 0.632 | 0.642 | 0.299 | 0.493 | 0.451 ± 0.177 | |
| R 2 | 3 | 0.763 | 0.702 | 0.730 | 0.200 | 0.534 | 0.390 | 0.553 |
| 6 | 0.777 | 0.817 | 0.812 | 0.265 | 0.569 | 0.433 | 0.612 | |
| 9 | 0.809 | 0.835 | 0.818 | 0.307 | 0.623 | 0.512 | 0.651 | |
| local | 0.848 | 0.854 | 0.860 | 0.368 | 0.644 | 0.534 | 0.685 | |
| AIC | 3 | 18.421 | 42.177 | 98.792 | 120.580 | 103.958 | 131.355 | 85.880 |
| 6 | 17.199 | 11.372 | 76.200 | 110.891 | 99.130 | 127.379 | 73.695 | |
| 9 | 10.095 | 6.847 | 74.243 | 106.892 | 87.499 | 119.566 | 67.524 | |
| local | -1.140 | -6.145 | 54.581 | 100.960 | 82.563 | 117.055 | 57.979 | |
| RMSE | 3 | 1.191 | 1.333 | 1.901 | 2.458 | 1.785 | 3.186 | 1.976 |
| 6 | 1.166 | 1.060 | 1.615 | 2.256 | 1.728 | 3.074 | 1.817 | |
| 9 | 1.084 | 1.012 | 1.591 | 2.190 | 1.617 | 2.858 | 1.725 | |
| local | 0.971 | 0.952 | 1.401 | 2.093 | 1.573 | 2.794 | 1.631 | |
e
i
|
3 | 0.921 | 1.022 | 1.530 | 1.889 | 1.417 | 2.461 | 1.540 |
| 6 | 0.893 | 0.789 | 1.288 | 1.808 | 1.394 | 2.376 | 1.425 | |
| 9 | 0.825 | 0.952 | 1.261 | 1.757 | 1.288 | 2.200 | 1.380 | |
| local | 0.768 | 0.698 | 1.074 | 1.636 | 1.251 | 2.176 | 1.267 | |
| σ ei | 3 | 7.003 | 9.597 | 6.695 | 11.070 | 15.330 | 14.798 | 10.749 |
| 6 | 7.500 | 6.420 | 8.304 | 10.867 | 17.376 | 17.215 | 11.280 | |
| 9 | 5.565 | 6.510 | 8.097 | 9.912 | 13.852 | 15.041 | 9.830 | |
| local | 5.794 | 6.611 | 9.888 | 14.152 | 12.311 | 17.593 | 11.058 | |
Classification and regression trees
In the CART model, all the selected variables were significant in the resulting regression tree, with a degree of significance of 100 for dbh, 97 for the mean diameter, 94 for stand age, and 57 for the site index. The resulting tree consists of 19 splitting nodes and 20 homogeneous groups (final nodes). Tab. 5 shows the actual values of the splitting criteria and the resulting values of regression tree, while Tab. 6 shows the goodness-of-fit statistics obtained for the resulting tree using the two data subsets (training and validation). The results correctly indicated that tree height increases with increasing stand age and dbh, supporting the use of CART models in different stages of the stand development. However, the achieved regression tree showed that trees from stands with lower mean diameter are taller than trees from stands with higher mean diameter, which seems biologically counter-intuitive. We therefore explored the characteristics of forest stands from which these trees originated, finding highly heterogeneous density values across forest stands. Indeed, competition for sunlight in high-density stands results in tall and slender trees, while more resources are allocated by trees to radial growth in low density-stands due to the lower crown competition. In our opinion, this may explain the observed anomaly.
Tab. 5 - Results of the regression tree construction. (LB): left branch; (RB): right branch; (N): number of trees in node; (μh): mean value of the height distribution; (SP): splitting variable; (dbh): diameter at breast height of a tree; (dbhm): mean diameter; (A): stand age; (SI): site index; (f): final nodes.
| Node | LB of the node |
RB of the node |
N | μh (m) | SP | Value of SP |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 860 | 27.97 | dbhm | 30.1 |
| 2 | 4 | 5 | 380 | 22.09 | A | 48 |
| 4 | 6 | 7 | 197 | 19.09 | dbh | 20.5 |
| 6 | 8 | 9 | 93 | 17.04 | dbh | 12.5 |
| 8f | - | - | 11 | 12.82 | - | - |
| 9 | 10 | 11 | 82 | 17.60 | dbh | 16.5 |
| 10f | - | - | 26 | 16.29 | - | - |
| 11 | 12 | 13 | 56 | 18.21 | A | 39 |
| 12f | - | - | 41 | 17.54 | - | - |
| 13f | - | - | 15 | 20.07 | - | - |
| 7 | 14 | 15 | 104 | 20.92 | A | 39 |
| 14f | - | - | 47 | 19.16 | - | - |
| 15f | - | - | 57 | 22.37 | - | - |
| 5 | 18 | 19 | 183 | 25.33 | dbh | 23.5 |
| 18 | 20 | 21 | 78 | 22.54 | dbh | 16.5 |
| 20f | - | - | 16 | 19.63 | - | - |
| 21f | - | - | 62 | 23.29 | - | - |
| 19 | 24 | 25 | 105 | 27.40 | SI | 32 |
| 24f | - | - | 46 | 29.01 | - | - |
| 25 | 26 | 27 | 59 | 26.14 | dbh | 30.5 |
| 26f | - | - | 42 | 25.62 | - | - |
| 27f | - | - | 17 | 27.44 | - | - |
| 3 | 28 | 29 | 480 | 32.62 | dbh | 32.5 |
| 28 | 30 | 31 | 126 | 28.99 | dbh | 23.5 |
| 30f | - | - | 16 | 24.56 | - | - |
| 31 | 32 | 33 | 110 | 29.64 | A | 75 |
| 32f | - | - | 23 | 27.85 | - | - |
| 33 | 34 | 35 | 87 | 30.11 | dbhm | 39.3 |
| 34f | - | - | 18 | 31.81 | - | - |
| 35f | - | - | 69 | 29.67 | - | - |
| 29 | 40 | 41 | 354 | 33.91 | dbh | 46.5 |
| 40 | 42 | 43 | 269 | 33.18 | A | 75 |
| 42f | - | - | 25 | 29.70 | - | - |
| 43 | 44 | 45 | 244 | 33.53 | dbhm | 39.3 |
| 44f | - | - | 45 | 35.52 | - | - |
| 45 | 46 | 47 | 199 | 33.08 | dbh | 40.5 |
| 46f | - | - | 111 | 32.51 | - | - |
| 47f | - | - | 88 | 33.80 | - | - |
| 41f | - | - | 85 | 36.22 | - | - |
Tab. 6 - Goodness-of-fit statistics obtained using the CART model. (R2): coefficient of determination; (ei): mean of residuals; (SEei ): standard error of residuals; (σei): residual standard deviation.
| Statistics | Training data | Validation data |
|---|---|---|
| R 2 | 0.940 | 0.804 |
| e i | 0.000 | -0.585 |
| SE ei | 0.054 | 0.102 |
| σ ei | 1.590 | 2.746 |
Further, based on the results from the CART model, trees from stands with lower site index were taller than those from stands with higher site index, which also seems illogical. A deeper analysis of the stands with lower site index revealed that the selected trees in these stands were mostly dominant and growing in better conditions than trees with mean diameter and height. Indeed, the site index in this study was not determined according to the height of dominant trees but to that of trees with mean height. The regression tree therefore correctly ranked these individuals into one homogeneous group based on more predictors than only site index (especially according to diameter at breast height). All the independent variables used in the CART model are thus biologically justified.
Comparison of OLS with LME and CART
The goodness-of-fit statistics obtained with each regression technique are compared in Tab. 7. The CART technique provided the lowest mean and standard deviation of residuals, but also a high average deviation of the fitted values from those predicted by the local model (in some cases close to 1 m). Although such deviation could still be considered acceptable for some applications (e.g., the use in volume tables for determining the standing volume), we consider CART modeling significantly biased as compared with the OLS method. Similar statistics were obtained by using the mixed model calibrated with only 3 tree heights in 3 diameter intervals. In both cases, small deviations could be observed in individual stands, but the average deviation of the predictions obtained by the two methods from those of the OLS model was high (Tab. 7). On the other hand, once a regression tree has already been established, its application to other stands does not require any additional height measurement. In our case, to estimate the height of a specific tree we need to know its diameter at breast height (to be measured) and the stand variables, such as mean diameter, stand age, and site index, which can be easily drawn from forest management plans.
Tab. 7 - Comparison of goodness-of-fit statistics for LME (mixed model), CART (regression tree model) and OLS (local model) techniques. (R2): coefficient of determination; (ei): mean of residuals; (σei): residual standard deviation; (Δi ): average deviation of the fitted values obtained using the LME model and the fitted values obtained using the OLS model.
| Model | Stats | Forest stand no. | Mean | |||||
|---|---|---|---|---|---|---|---|---|
| 3 | 5 | 11 | 14 | 19 | 22 | |||
| LME | R 2 | 0.809 | 0.835 | 0.818 | 0.307 | 0.623 | 0.512 | 0.651 |
| e i | 0.825 | 0.952 | 1.261 | 1.757 | 1.288 | 2.200 | 1.380 | |
| σ ei | 5.565 | 6.510 | 8.097 | 9.912 | 13.852 | 15.041 | 9.830 | |
| Δ i | 0.400 | 0.239 | 0.632 | 0.642 | 0.299 | 0.493 | 0.451 | |
| CART | R 2 | 0.783 | 0.727 | 0.767 | 0.202 | 0.570 | 0.465 | 0.586 |
| e i | 0.089 | 0.094 | 0.148 | -0.530 | -0.021 | 0.144 | -0.013 | |
| σ ei | 1.146 | 1.293 | 1.788 | 2.272 | 1.718 | 2.963 | 1.863 | |
| Δ i | 3.546 | 2.303 | 2.220 | 1.258 | 1.179 | 1.216 | 1.954 | |
| OLS | R 2 | 0.848 | 0.854 | 0.860 | 0.368 | 0.644 | 0.534 | 0.685 |
| e i | 0.768 | 0.698 | 1.074 | 1.636 | 1.251 | 2.176 | 1.267 | |
| σ ei | 5.794 | 6.611 | 9.888 | 14.152 | 12.311 | 17.593 | 11.058 | |
The LME technique provided good parameter estimates of the residual distribution, though their values were higher than those of the CART model. However, its predictions were very close to those achieved with the local model using the OLS method (Tab. 7), and in some cases even better. The average deviation of predictions by both the local model and the mixed model (based on 9 height measurements) were small both in the individual forest stands and overall. We concluded that the LME technique is superior to the CART technique for the prediction of height based on dbh, and that the mixed model can replace the local model without significant losses of goodness-of-fit of the height curve model. Moreover, the mixed model also guarantees a consisting reduction in the variability of residuals.
Discussion
The results of this study revealed that tree height can be accurately predicted based on diameter at the breast height by including the stand age and its logarithmic transformation as random factors in the mixed model considered. Moreover, the mixed model removed the heteroscedasticity occurring in the global model (Fig. 1), as previously reported by Drápela ([13]). However, the inclusion of stand age as a stand variable makes the use of the above model suitable only to even-aged forests. Mixed models for height prediction were previously applied to even-aged forests, but using different stand variables, such as the dominant height and dominant diameter ([10]), the mean diameter ([37]), or a combination of stand variables ([38], [2]). Other authors have focused on uneven-aged forest using the mean/dominant diameter or the dominant height as predictors ([9]), as the use of stand age is unfeasible. Eerikäinen ([14]) stated that the inclusion of the dominant height in models may reflect the site index as an indicator of the production potential of a site. All the above evidences support the hypothesis that the inclusion of stand variables into the model improves its final accuracy ([41]).
The applicability of the proposed mixed model in practice has been assessed by comparing the effect of different ways of selecting trees for height measurement to be used in the calibrated conditional prediction. Using the first method, 1 to 5 trees within ± 2 cm from the mean diameter were selected, while the second was based on 3, 6, or 9 trees randomly selected within 4 cm intervals from the thinnest, medium and thickest trees. Other papers also included two ways of tree selection, namely random selection of different number of trees and a selection focused only on a portion of the diameter range. For example, random selection of trees has been applied by Adame et al. ([2]) and Kangas & Haara ([21]), who selected 1 to 4 trees, while Trincado et al. ([42]) chose only 1 tree. Sharma & Parton ([38]) and Schmidt et al. ([37]) carried out a selection of 1 to 3 trees. Crecente-Campo et al. ([10]) and Castaño-Santamaría et al. ([9]) randomly selected 1 to 10 trees in each of three ranges of minimum, mean and largest diameters, as well as 3, 6, or 9 trees in several ranges of the smallest, largest and mean diameter.
Calibration results showed that random selection of trees close to the mean diameter is not feasible, as it provides predictions too biased compared to local models or not biologically justified (height decreasing as the diameter increases). Crecente-Campo et al. ([10]) reported similar results, with very high mean and standard error of residuals of the resulting model using 1 to 10 trees of mean diameter. Castaño-Santamaría et al. ([9]) using trees of mean diameter did not find such high means of residuals, but recommended the use of trees with smaller diameter to achieve better results in the calibrated conditional prediction.
The second method, based on the selection of trees from more diameter intervals, proved to yield better results in this study. Similarly, Crecente-Campo et al. ([10]) and Castaño-Santamaría et al. ([9]) reported that this approach improves the predictive power of the model. The results of this study confirm that the goodness-of-fit of the model increases by increasing the number of measured trees, in accordance with Trincado et al. ([42]), Adame et al. ([2]) and Kangas & Haara ([21]).
Sharma & Parton ([38]) and Schmidt et al. ([37]) compared both LME model with fixed effects marginal prediction and calibrated conditional prediction, concluding that the latter provides better results even when only one tree is used for model calibration.
Van Laar & Akça ([43]) recommended a minimum number of 20-25 tree height measurements for determining a local model of the height curve, to be used for example for calculation of the standing volume using the volume tables. We found that the reduction in number (up to 6 or 9) of the required measured heights through the calibrated model did not invalidate the model predictability, but at the same time ensures a reduction in costs and time of data collection. According to Crecente-Campo et al. ([10]), this reduction in the number of required measurements makes the application of mixed models highly effective and useful.
The results obtained using the CART model indicated that tree height depends on the diameter at breast height, mean diameter, stand age, and site index. Strongly nonlinear relationships exist between the height and the above variables. According to Gómez et al. ([18]), the CART method should be preferred to parametric models as it is more suitable to analyze nonlinear relationships as well as data with hierachical structure ([11], [39]).
Indeed, the strong dependence among independent variables (e.g., dbh and age) could bring about problems of multicollinearity when parametric methods are used. Contrastingly, CART models have been reported to resist to multicollinearity ([39]), as CART considers the high-hierarchy variables to be more important than low-hierarchy variables, and the independent variables to have greater influence on the results than their mutual correlations. Consequently, there are no biased estimates of parameters ([3]). Further, the easy description of interactions between variables make this method a useful tool for the interpretation of the results, particularly when the number of independent variables increases and other model types are very difficult to interpret ([11]).
Our results showed that tree height can be quite easily predicted using the CART method. CART models have been previously applied to estimate different variables in forestry, such as tree mortality ([12], [15]), site index ([3]) and the form factor of sample trees ([33]). The interpretation of the regression tree obtained with the CART model is very easy and intuitive, which makes this method a suitable tool for classifying forest stands ([18]). Moreover, once the regression tree is achieved, no additional measurements are required, thus reducing the costs of data collection.
In this study, the comparison of mixed model, regression tree, and the local model indicated that LME performed better than CART in predicting tree height based on dbh. The CART method also provided results significantly worse than those obtained with the OLS method (local model), except for parameters of the residual distribution. Moisen & Frescino ([29]) compared several modeling techniques to predict various forest characteristics, confirming the worse performances of CART compared to OLS methods. Aertsen et al. ([3]) compared CART with multiple linear regression (MLR), generalized additive models (GAM) boosted regression trees (BRT) and artificial neural networks (ANN), confirming that CART is more user-friendly than the other four modeling techniques.
Regarding the comparison between LME and OLS models, we found that predictions obtained with LME only slightly differs from those of the local model. Crecente-Campo et al. ([10]) found that calibrated LME provides significantly better results than the global model, thanks to the calibrated conditional prediction of the local model. Similar results were achieved by Castaño-Santamaría et al. ([9]). We concluded that the mixed model with calibrated conditional prediction is best suited for the regionalization of the global model, e.g., up to the level of the forest stand. We recommend to calibrate the model based on 6 to 9 tree height measurements in a given stand, depending on the required level of accuracy.
Acknowledgements
The authors thank two anonymous reviewers whose comments helped to improve an earlier version of the manuscript. We would also like to thank Jan Kadavý for his valuable comments.
References
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Karel Drápela
Department of Forest Management and Applied Geoinformatics, Faculty of Forestry and Wood Technology, Mendel University in Brno, Brno, 613 00 (Czech Republic)
Corresponding author
Paper Info
Citation
Adamec Z, Drápela K (2016). Comparison of parametric and nonparametric methods for modeling height-diameter relationships. iForest 10: 1-8. - doi: 10.3832/ifor1928-009
Academic Editor
Piermaria Corona
Paper history
Received: Nov 24, 2015
Accepted: Jul 07, 2016
First online: Oct 19, 2016
Publication Date: Feb 28, 2017
Publication Time: 3.47 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2016
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 49558
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 41344
Abstract Page Views: 2830
PDF Downloads: 3981
Citation/Reference Downloads: 49
XML Downloads: 1354
Web Metrics
Days since publication: 3420
Overall contacts: 49558
Avg. contacts per week: 101.43
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2017): 12
Average cites per year: 1.33
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Estimating biomass of mixed and uneven-aged forests using spectral data and a hybrid model combining regression trees and linear models
vol. 9, pp. 226-234 (online: 21 September 2015)
Research Articles
Bayesian geographically weighted regression and its application for local modeling of relationships between tree variables
vol. 11, pp. 542-552 (online: 01 September 2018)
Research Articles
A geographically weighted deep neural network model for research on the spatial distribution of the down dead wood volume in Liangshui National Nature Reserve (China)
vol. 14, pp. 353-361 (online: 27 July 2021)
Research Articles
Modeling compatible taper and stem volume of pure Scots pine stands in Northeastern Turkey
vol. 16, pp. 38-46 (online: 22 January 2023)
Research Articles
Modeling stand mortality using Poisson mixture models with mixed-effects
vol. 8, pp. 333-338 (online: 05 September 2014)
Research Articles
Nonlinear mixed model approaches to estimating merchantable bole volume for Pinus occidentalis
vol. 5, pp. 247-254 (online: 24 October 2012)
Research Articles
The patterns of nearest neighbor trees in a temperate forest
vol. 15, pp. 315-321 (online: 23 August 2022)
Research Articles
Classification and mapping of Spanish Mediterranean mixed forests
vol. 12, pp. 480-487 (online: 14 October 2019)
Research Articles
Distribution factors of the epiphytic lichen Lobaria pulmonaria (L.) Hoffm. at local and regional spatial scales in the Caucasus: combining species distribution modelling and ecological niche theory
vol. 17, pp. 120-131 (online: 30 April 2024)
Research Articles
Estimation of stand crown cover using a generalized crown diameter model: application for the analysis of Portuguese cork oak stands stocking evolution
vol. 9, pp. 437-444 (online: 02 December 2015)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword