Patterns of genetic diversity in European beech (Fagus sylvatica L.) at the eastern margins of its distribution range
Elena Ciocîrlan (1), Nicolae Sofletea (1), Fulvio Ducci (2), Alexandru Lucian Curtu (1)
iForest - Biogeosciences and Forestry, Volume 10, Issue 6, Pages 916-922 (2017)
doi: https://doi.org/10.3832/ifor2446-010
Published: Dec 10, 2017 - Copyright © 2017 SISEF
Research Articles
Collection/Special Issue: COST Action FP1202
Strengthening conservation: a key issue for adaptation of marginal/peripheral populations of forest trees to climate change in Europe (MaP-FGR)
Guest Editors: Fulvio Ducci, Kevin Donnelly
Abstract
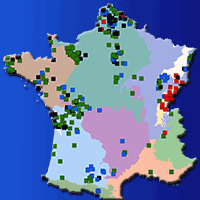
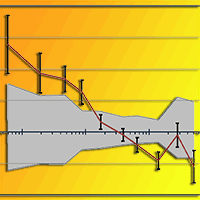
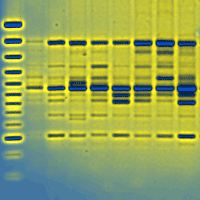
Populations located at the periphery of the species’ distribution range may play an important role in the context of climate change. These peripheral populations may contain specific adaptations as a result of extreme environmental conditions. The aim of this paper was to assess within population genetic diversity and among population differentiation in one of the most important forest tree species in Europe, European beech (Fagus sylvatica), at the eastern margins of its natural range. We analysed four peripheral, isolated populations and five core populations from the continuous natural range along the Carpathian Mountains using a set of microsatellite markers. Higher levels of genetic diversity as measured by allelic richness (7.34 vs. 6.50) and observed heterozygosity (0.71 vs. 0.59) were detected in core populations than in peripheral ones. Population differentiation was slightly higher among peripheral populations than among core, Carpathian populations. There was strong evidence of bottleneck effects in two out of the four peripheral, isolated populations. Both core, Carpathian populations and peripheral, lowlands populations share the same chloroplast haplotype suggesting a common geographical origin from the putative Moravian refuge area. Past long distance founding events with material from the Carpathian mountain chain might explain the occurrence of small, isolated beech populations towards the steppe in the south-east of Romania. Our genetic data may contribute to a better understanding of the evolutionary history of the remnants of beech scattered occurrences at the eastern margins of species’ distribution range.
Keywords
Fagus sylvatica, Genetic Diversity, Peripheral Populations, Bottleneck Effect
Authors’ Info
Authors’ address
Nicolae Sofletea
Alexandru Lucian Curtu
Department of Forest Sciences, Transilvania University of Brasov, Sirul Beethoven-1, 500123 Brasov (Romania)
CREA SEL, Consiglio per la ricerca in agricoltura e l’analisi dell’economia agraria, Forestry Research Centre, I-52100 Arezzo (Italy)
Corresponding author
Paper Info
Citation
Ciocîrlan E, Sofletea N, Ducci F, Curtu AL (2017). Patterns of genetic diversity in European beech (Fagus sylvatica L.) at the eastern margins of its distribution range. iForest 10: 916-922. - doi: 10.3832/ifor2446-010
Academic Editor
Fulvio Ducci
Paper history
Received: Mar 29, 2017
Accepted: Aug 23, 2017
First online: Dec 10, 2017
Publication Date: Dec 31, 2017
Publication Time: 3.63 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2017
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 51819
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 42845
Abstract Page Views: 3692
PDF Downloads: 4059
Citation/Reference Downloads: 23
XML Downloads: 1200
Web Metrics
Days since publication: 2985
Overall contacts: 51819
Avg. contacts per week: 121.52
Citation Metrics
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2017): 10
Average cites per year: 1.11
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
References
Fagetele primare din România, o contributie la Patrimoniul Mondial UNESCO [Romania’s primary beech forests, a contribution to UNESCO World Heritage]. Bucovina Forestiera 14: 77-85. [in Romanian]
Gscholar
Comparative morphological analyses in marginal beech populations. Bulletin of the Transilvania University of Brasov, Forestry, Wood Industry, Agricultural Food Engineering, Series II 7 (56): 7-12.
Gscholar
Valoarea documentara fitoistorica a mlastinii de turba de la Mangalia-Herghelie (Jud. Constanta) [The phyto-historical documentary value of the peat bog from Mangalia-Herghelie (Constanta County)]. Contributii Botanice, Gradina Botanica “Alexandru Borza”, Cluj-Napoca, Romania, pp. 41-53. [in Romanian]
Gscholar
Pozitia sistematica si origina fagilor dela Luncavita (Dobrogea de Nord) [The systematic position and origin of the beech trees from Luncavita (Northern Dobrogea)]. Revista Padurilor 1-2: 25-31. [in Romanian]
Gscholar
A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genomics 11 (1): 570.
CrossRef | Gscholar
Zonele de vegetatie lemnoasa din România [The woody vegetation zones of Romania]. Editura Cartea Româneasca, Bucuresti, Romania, pp. 268. [in Romanian]
Gscholar
Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. Forest Ecology and Management 375: 66-75.
CrossRef | Gscholar
Prezenta fagului în padurile Ocolului Silvic Snagov [The presence of beech in forests of Snagov Forest District]. Revista Padurilor 88: 367-370. [in Romanian]
Gscholar
State of Europe’s Forests 2011. Status and Trends in Sustainable Forest Management in Europe. Ministerial Conference on the Protection of Forests in Europe, FOREST EUROPE Liaison Unit, Oslo, Norway, pp. 344.
Gscholar
Insulele de fag din Dobrogea [Beech islands of Dobrogea]. Revista Padurilor 4: 231-242. [in Romanian]
Gscholar
A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytologist 171: 199-221.
CrossRef | Gscholar
Molecular evolutionary genetics. Columbia University Press, New York, USA, pp. 512.
Gscholar
The vegetation of the natural reserve Valea Fagilor-Luncavita (Tulcea County, Romania). Contributii Botanice, Gradina Botanica “Alexandru Borza”, Cluj-Napoca XLVI: 17-32.
Gscholar
ARLEQUIN ver 2.000. A software for population genetics data analysis. Genetics and Biometry Laboratory, Dept. of Anthropology and Ecology, University of Geneva, Geneva, Switzerland.
Gscholar
Flora forestiera lemnoasa a Romaniei [Woody forest flora of Romania]. Editura Ceres, Bucuresti, Romania, pp. 451. [in Romanian]
Gscholar