Monitoring of the incidence of Dutch Elm Disease and mortality in experimental plantations of French Ulmus minor clones
iForest - Biogeosciences and Forestry, Volume 15, Issue 4, Pages 289-298 (2022)
doi: https://doi.org/10.3832/ifor3820-015
Published: Jul 29, 2022 - Copyright © 2022 SISEF
Research Articles
Abstract
The results of 16 experimental plantations of Ulmus minor clones of the French national collection are presented as a complement to a previous work (Collin et al. 2020) on the outcome of the French Programme for the Conservation of Native Elm Genetic Resources. A total of 710 elms from 38 clones were planted in three different regions of France using two types of experimental designs and exposed to natural infection by Dutch Elm Disease (DED). DED infection and subsequent mortality were monitored in rectangular monospecific plantations (“plot-tests”), comprising at least 100 elms (10 clones). Linear plantations of 15 to 36 elms intermixed with other trees and shrubs (“hedge-tests”) investigated the feasibility of using native field elm clones in hedge reconstruction projects. After at least 9 (up to 17) years of experimentation, overall DED infection frequency was 33%, with scores above 63% in the oldest plantation and in two fast-growing tests. The overall mortality in the 232 diseased trees was 21%, reaching 29% in the oldest plantation and 64% in a hedge-test on high quality soil, suggesting a possible effect of the very fast growth of the trees. A few clones showed an interesting lower infection frequency or some ability to recover, whereas some others were found quickly infected in several tests and could serve in future experiments on clone attractiveness for DED vectors. Practical conclusions for genetic resources conservation consist of recommendations for the establishment of conservation plantations using regional clones.
Keywords
Ulmus minor, Plantation, Dutch Elm Disease, Ophiostoma novo-ulmi, Genetic Resources, France
Introduction
The second pandemic of Dutch Elm Disease (DED) that swept Western Europe in the 1970s ([4], [5]) raised interest in the conservation of native elm genetic resources ([17]) and the hope that clones of old trees having survived the epidemic might hold a higher tolerance to DED ([24]). Collin et al. ([8]) have presented the results of the French National Programme for the Conservation of Native Elm Genetic Resources launched in 1987. This programme was initially focused on the ex-situ conservation of clones of adult field elm (Ulmus minor Mill.) survivors of the DED pandemic. It was later expanded to include the in-situ dynamic conservation of populations of European white elm (U. laevis Pall.) and wych elm (U. glabra Huds.). The national collection contains 441 clones, partly characterized and evaluated in a European project. Collin et al. ([8]) give a brief summary of the overall sanitary status of the experimental plantations set up by CEMAGREF (renamed IRSTEA in 2012 and INRAE after merging with INRA in 2020) but no details of the tests and the clones tested. Such details are given in the present study.
Unlike its Spanish counterpart ([17]), the French elm conservation programme was not linked to a breeding programme and did not involve quantitative genetics ([27]) or genomics approaches. Artificial inoculation tests were carried out in nurseries to evaluate the susceptibility of many clones to the fungal agent of DED Ophiostoma novo-ulmi Brasier. Because no native elm clone reached the high levels of resistance of Eurasian hybrid cultivars such as LUTECE® ’Nanguen’, their cultivation in pure stands was not recommended ([22], [23]) but, as some of them showed a strong recovery ability one year after inoculation, it seemed possible to use them in multi-species plantations, e.g., for the reconstruction of countryside hedges ([22], [26]). However, artificial inoculation tests carried out on four-year-old plants do not produce a full picture of the long-term response of these elms to O. novo-ulmi. Thus, experimental plantations were felt necessary for three main reasons: (i) to better evaluate the frequency and durability of recovery episodes (or contrariwise, of disease recurrence) which cannot be monitored in the short time-span of a nursery test; (ii) to observe the tolerance performance of different clones submitted to the variation of the DED pathosystem under diverse natural conditions; (iii) to provide tree planters and conservationists with practical information of interest for hedge reconstruction and the dynamic conservation of elm genetic resources.
The first reason came from the experience gained in the inoculation tests conducted by INRA and IRSTEA, where many trees neither succumbed nor recovered but suffered stem dieback and/or leaf wilt of different magnitudes and dynamics. Some clones with trees severely wilted in the inoculation year were able to improve in the second year and sometimes turned asymptomatic for several years, whereas some clones with lighter wilt in the inoculation year showed recurrent wilt in the second and third years. More years of observations are needed to see if recoveries can be permanent, but nursery plantations have to be uprooted two or three years after inoculation, i.e., before trees are excessively large and spacing becomes insufficient. The second reason came from the need to complement the missing part of tolerance evaluation in artificial inoculations tests, which ignore the role of DED vectors, and particularly the possible differences between elm clones in attractiveness for bark beetles. As reviewed by Martin et al. ([17]), interactions between actors of the pathosystem (including the tree microbiota, and viruses deleterious to the pathogenic fungus) are complex, not to mention the role of edaphic and climatic factors. Thirdly, experimental plantations are absolutely required to observe tree survival and vigour in the long term, as well as clone shape and ornamental features (leaf aspect, corky ridges, etc.), age of sexual maturity and other characters of practical interest for hedge planters and conservationists. Results for this third objective of experimental plantations are to be presented in a separate article (Collin et al., in preparation).
To address these needs, IRSTEA established two types of experimental plantations exposing elm clones to natural infection by DED. These experiments aimed at observing the long-term field performance and DED tolerance of a broad array of field elm clones belonging to the national collection. In addition, the hedgerow-type plantations were evaluated for the capacity of native elms to thrive in hedge restoration projects with other trees and shrubs. These objectives were met, as the results presented here were extracted from data based on more than 700 elm trees from 38 native field elm clones, after nine years (or more) of experimentation in 16 tests in three different regions of France and in different environments. Similar information about native European elm clones has rarely been published. Girard ([11]) reports on 11 plantations of the Institut pour le Développement Forestier (IDF), comparing five elm cultivars and four native elm clones of the French national collection but only four years after plantation, i.e., before infection by DED, except in one test.
Because the objectives of the experimental plantations were not focused on a specific research question or on a small number of superior clones, some of the results presented here are mostly descriptive but their publication has been encouraged by pathologists and forest genetic resources conservationists interested in the disease progress and mortality rate of naturally infected elms. These results can also provide a solid foundation for future research based on field elm clones showing contrasted levels of infection rate or of recovery ability. Moreover, the maps and data files supplied as Supplementary material to this article offer the possibility for a long-term re-assessment of the three most interesting experimental plantations.
Material and methods
Material
With 4 exceptions, the 38 clones considered in this study were U. minor clones (Tab. 1). Species assignment was based on leaf and bud characters and confirmed by molecular markers (RAPDs and ISSRs) when available ([12]). Four clones of intermediate forms with the wych elm were added because they belong to the natural elmscape of Basse-Normandie, the original region of many clones and the place of several experimental plantations. The clonal codes of those putative hybrids or backcrosses (Ulmus × hollandica Mill.) have a special suffix (“mg” or “gm” ending) indicating their hybrid nature and the prominent parental traits of their leaf and bud characters, e.g., “mg” looking more like a field elm but with some features of the wych elm. The “*” ending of F352 clone code indicates that species assignment needs to be checked with molecular markers for possible introgression by an introduced elm species.
Tab. 1 - Number of plants included in the study per clone and per plantation. In the “clone” column, codes with “mg” or “gm” endings indicate putative hybrids or backcrosses with U. glabra. (*): species assignment to be checked for possible introgression by an introduced elm species. (IT): value in inoculation tests ([22] and comparison with cv. ’Christine Buisman’ in a test conducted by the authors in 2013); (+): among the less susceptible clones; (-): among the more susceptible clones; (?): untested or unclear results.
| test/clone | Clone origin | IT | 11 | 12 | 13 | 14 | 21 | 22 | 23 | 24 | 25 | 26 | 31 | 32 | 33 | 34 | 35 | 36 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F020 | Basse-Normandie | + | - | - | - | - | 8 | - | - | - | - | - | 9 | - | - | - | - | - | 17 |
| F024 | Basse-Normandie | + | - | - | - | - | 9 | - | - | - | - | - | 9 | - | - | - | - | - | 18 |
| F028 | Basse-Normandie | + | - | - | - | - | 10 | - | 6 | 6 | - | - | 9 | - | - | - | - | - | 31 |
| F032mg | Basse-Normandie | + | 11 | 14 | 4 | 10 | 9 | 6 | 5 | 6 | - | - | 10 | - | - | 5 | 4 | 5 | 89 |
| F034 | Basse-Normandie | + | 4 | - | - | - | 10 | - | - | - | - | - | 7 | 5 | 5 | - | - | - | 31 |
| F035 | Basse-Normandie | + | - | - | - | - | - | - | - | 6 | - | - | 8 | - | - | - | - | - | 14 |
| F040 | Basse-Normandie | - | 7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 7 |
| F041gm | Basse-Normandie | + | - | - | - | 7 | - | - | - | - | - | - | 8 | - | - | - | 5 | - | 20 |
| F043 | Basse-Normandie | ? | - | - | - | 8 | - | - | - | - | - | - | - | - | - | - | - | - | 8 |
| F044 | Basse-Normandie | + | - | - | - | - | 8 | - | - | - | - | - | 6 | - | - | - | - | - | 14 |
| F050 | Basse-Normandie | ? | - | - | - | - | 9 | - | - | - | - | - | 9 | - | - | - | - | - | 18 |
| F052 | Basse-Normandie | + | 3 | - | - | 7 | - | - | - | - | - | - | - | - | - | - | - | - | 10 |
| F054 | Basse-Normandie | ? | - | - | - | - | - | - | - | - | 6 | - | - | - | - | - | - | - | 6 |
| F055 | Basse-Normandie | ? | 9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 9 |
| F056 | Basse-Normandie | + | - | - | - | - | 7 | - | - | - | - | - | 9 | - | - | - | - | - | 16 |
| F060 | Basse-Normandie | + | 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 8 |
| F071gm | Basse-Normandie | - | 7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 7 |
| F072 | Basse-Normandie | + | - | - | - | 9 | 9 | - | - | - | - | - | 5 | - | - | - | - | - | 23 |
| F077 | Basse-Normandie | + | 3 | - | - | 9 | - | - | - | - | - | - | 7 | - | - | - | - | - | 19 |
| F083 | Basse-Normandie | + | 12 | - | 5 | 10 | 10 | - | - | 6 | - | - | 9 | - | - | 5 | 5 | - | 62 |
| F085 | Basse-Normandie | - | - | - | - | 9 | - | - | - | - | - | - | - | - | - | - | - | - | 9 |
| F115 | Poitou-Charentes | + | - | - | - | - | 5 | - | - | - | 6 | 4 | 3 | - | - | - | - | 5 | 23 |
| F124mg | Poitou-Charentes | + | 7 | - | - | - | - | - | - | - | - | - | 7 | - | - | - | - | - | 14 |
| F133 | Poitou-Charentes | ? | - | - | - | - | - | - | - | - | - | 5 | - | - | 5 | - | - | - | 10 |
| F136 | Poitou-Charentes | ? | - | - | - | - | - | - | - | - | - | - | - | - | - | 5 | - | - | 5 |
| F140 | Poitou-Charentes | - | 11 | - | - | - | 10 | - | - | - | - | - | 10 | - | - | - | - | - | 31 |
| F144 | Poitou-Charentes | + | - | - | - | - | - | - | - | 4 | 6 | - | 7 | 4 | 6 | - | - | - | 27 |
| F147 | Poitou-Charentes | ? | - | - | - | - | - | - | - | - | - | 5 | - | - | - | 5 | - | 5 | 15 |
| F178 | Ile-de-France | ? | 6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 6 |
| F203 | Bretagne | ? | - | 6 | - | - | - | - | - | - | - | - | 8 | - | - | - | - | - | 14 |
| F205 | Bretagne | + | - | 9 | 2 | - | - | - | - | - | - | - | - | - | - | - | - | - | 11 |
| F250 | Picardie | - | - | 14 | 3 | 10 | - | - | - | - | - | - | - | - | - | - | - | - | 27 |
| F350 | Bretagne | + | - | - | - | - | - | 6 | 5 | - | 6 | - | 7 | 5 | - | - | - | - | 29 |
| F351 | Bretagne | + | - | 14 | 2 | - | - | - | - | - | - | - | - | - | - | 5 | - | - | 21 |
| F352* | Bretagne | + | - | - | - | - | - | - | - | 6 | - | - | 8 | - | - | - | - | - | 14 |
| F355 | Bretagne | ? | - | - | - | - | - | 6 | - | 2 | - | - | 6 | - | - | - | - | - | 14 |
| F393 | Bretagne | + | - | - | - | - | - | - | - | - | - | - | - | - | - | 5 | - | - | 5 |
| F471 | Rhône-Alpes | ? | - | - | - | 8 | - | - | - | - | - | - | - | - | - | - | - | - | 8 |
| Total | - | - | 88 | 57 | 16 | 87 | 104 | 18 | 16 | 36 | 24 | 14 | 161 | 14 | 16 | 30 | 14 | 15 | 710 |
| No. clones | - | - | 12 | 5 | 5 | 10 | 12 | 3 | 3 | 7 | 4 | 3 | 21 | 3 | 3 | 6 | 3 | 3 | 38 |
Twenty-two of the 38 clones considered here have shown some tolerance to O. novo-ulmi in artificial inoculation tests. Always severely wilted (>50%) in the inoculation year, they were far from reaching the high levels of resistance of Eurasian hybrid cultivars such as ’Lobel’ and LUTECE® ’Nanguen’, obtained in the Dutch breeding programme ([13]) and showing less than 15% wilt or no symptoms at all ([22], [26]). In an inoculation test carried out by the authors in 2013 (Collin & Rondouin, unpublished data), field elm clones F028 and F041gm performed nearly at the level of ’Christine Buisman’, a Spanish field elm clone with a moderate tolerance to DED selected in the Netherlands in 1936 and with trees still alive in the 2010s in plantations dating back to the 1950s (Heybroek HM, pers. comm.). The wilt percentages of ’Christine Buisman’, F028 and F041gm were respectively 54.5%, 64% and 69%. None of the inoculated trees of ’Christine Buisman’ and F041gm showed recurrent symptoms in the second year, and only two out of ten of F028 trees did so.
The 22 field elm clones with frequent recovery or limited symptoms (wilt <10%) in the second year of an inoculation test (as indicated in Tab. VIII of [22]), were classified in category “+” in column IT (i.e., “Inoculation test”) of Tab. 1. The 11 clones classified in category “?” were used in the experimental plantations because they were available in nursery and originated from the planting regions but their susceptibility to DED had not been tested. Only five clones belong to category “-”, indicative of high mortality and/or persistent severe wilt in inoculation tests. These five clones were planted in the experimental plantations either deliberately as susceptible control (F085) in one large test or involuntarily because their susceptibility was unknown or not firmly assessed at the time of their plantation.
The plant material was propagated by softwood cuttings ([20]) at the nursery of the Pôle National des Ressources Génétiques Forestières (PNRGF) of the Office National des Forêts (ONF) in Guémené-Penfao (Loire-Atlantique). After transplantation for one year in the nursery, the elms were sent for planting-out as bare-root plants. Plant height was above 1 m for the most vigorous clones, and between 0.7 and 1 m for the others.
Tab. 1 lists the 710 plants considered in this study. Plants damaged (by deer, forestry mulcher or wind, for examples) and plants less than 3 m high (abnormally weak and generally unattractive to bark beetles) were removed to preclude bias in the analysis. Clones represented by fewer than five undamaged plants higher than 3 m were also removed. Most of the 38 clones considered here were tested in only one or two plantations but a subset of 15 clones were tested in at least three plantations, the maximum being 12 plantations for clone F032mg and eight for F083.
Methods
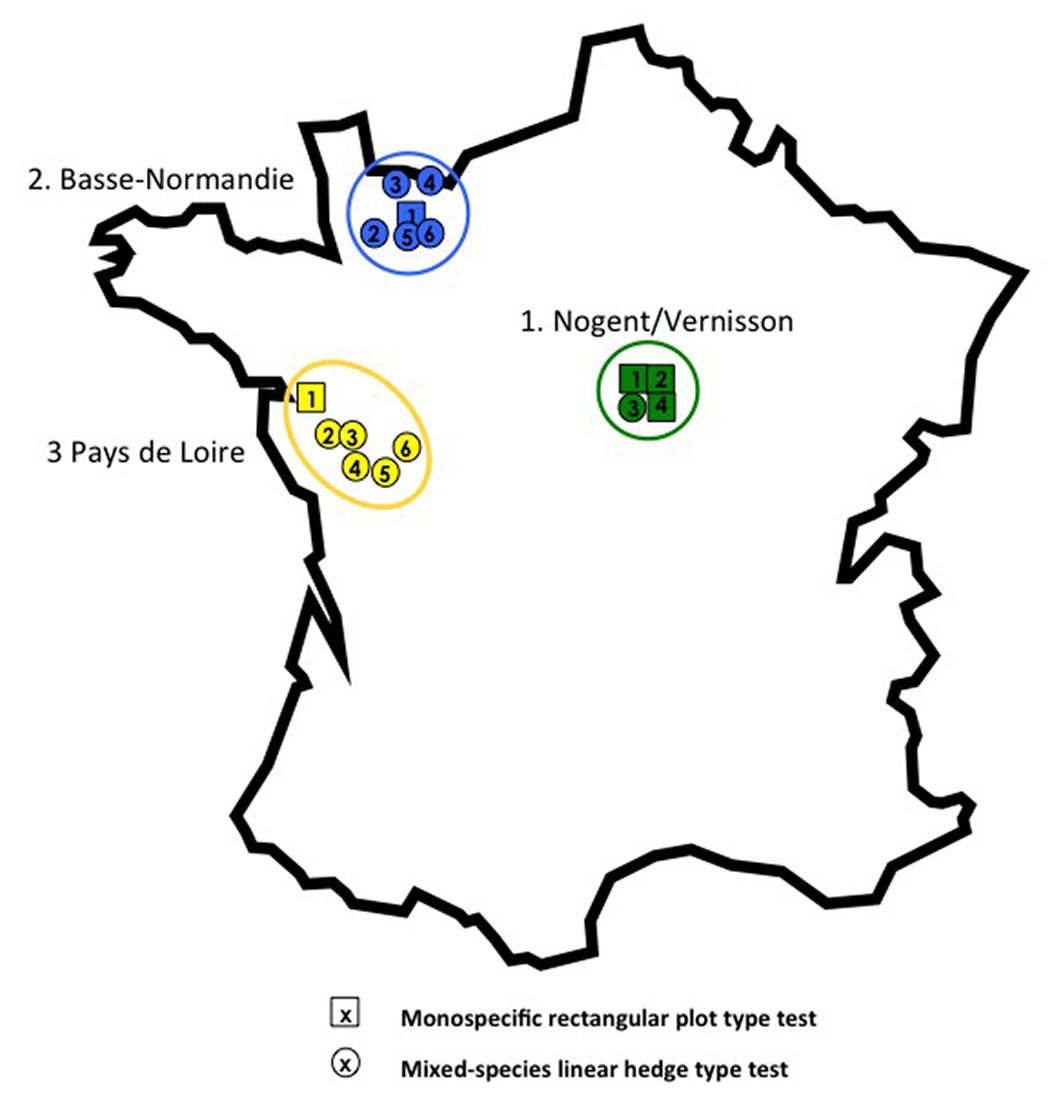
The elm experimental plantations of IRSTEA considered in this study (Tab. 2) were planted between 2002 and 2011. They were established with numerous partners, mainly in western France (Fig. 1). In the “plot” type monospecific plantations, the elms were planted about 5 m apart on the line and between the lines (Fig. S3.1-S3.3 in Supplementary material). In the “hedge” type plantations, they were planted about 6 to 8 m apart on a single line, in association with other tree and shrub species. In both cases, the experimental protocol ensured an equal and randomized distribution of clones throughout the plantation, with at least five plants per clone in the hedges, and ten in the plots. All plantations followed the single-tree plot design, except the two oldest (2002 and 2005), composed of square experimental units of 4 plants × 3 blocks. The latter design was used for the first tests because it was valued by elm breeders for the long-term evaluation of their best hybrid cultivars. However, it proved less suitable for the evaluation of DED-susceptible material suffering severe dieback and mortality at young age. A single-tree plot design was preferred here because it has advantages when the number of trees per clone is low and because it precludes the risk that four plants of the same clone are infected just by a local effect of the vectors’ flight or by root grafts with an infected neighbouring tree of the same clone. In hedges, the protocol ensured that all elms would be planted next to the same shrub species (e.g., Viburnum lantana on the left side, Cornus sanguinea on the right) to prevent any difference in neighbourhood effect.
Tab. 2 - Plantations characteristics. The first figure of the test code number indicates the region of plantation as shown on Fig. 1. (*): number of plants (damaged excluded) ; (**): number of clones of Ulmus minor or U. x hollandica. (***): age (number of growth seasons) at the time of the last assessment (generally summer of 2018, except for tests #14 and #31, assessed in summer of 2019).
| Test code |
Community (locality) | Lat N / Long E | Plantation type |
Previous land use |
1st year |
No. plants* |
No. clones** |
Age*** |
|---|---|---|---|---|---|---|---|---|
| 11 | Nogent/V. (Toussaint) | 47.846719 / 2.76256 | plot | woodland | 2002 | 88 | 12 | 17 |
| 12 | Nogent/V. (Montagne) | 47.844536 / 2.75552 | plot | woodland | 2005 | 57 | 5 | 14 |
| 13 | Nogent/V. (Chem. Talonet) | 47.838275 / 2.76518 | hedge | old hedge | 2006 | 16 | 5 | 13 |
| 14 | Nogent/V. (Le Talonet) | 47.840248 / 2.76301 | plot | woodland | 2008 | 87 | 10 | 12 |
| 21 | Banneville/Ajon (plot) | 49.069657 / -0.55974 | plot | hayfield | 2007 | 104 | 12 | 12 |
| 22 | Guilberville | 48.958610 / -0.97673 | hedge | crop field | 2007 | 18 | 3 | 12 |
| 23 | Saint-Gabriel-Brecy | 49.279630 / -0.56378 | hedge | crop field | 2007 | 16 | 3 | 12 |
| 24 | Courseulles-sur-Mer | 49.314853 / -0.46424 | hedge | crop field | 2007 | 36 | 7 | 12 |
| 25 | Banneville/Ajon (hedge 1) | 49.070220 / -0.55970 | hedge | hayfield | 2007 | 24 | 4 | 12 |
| 26 | Banneville/Ajon (hedge 2) | 49.069474 / -0.55923 | hedge | hayfield | 2007 | 14 | 3 | 12 |
| 31 | Saint-Herblain | 47.255806 / -1.63737 | plot | pasture | 2007 | 161 | 21 | 13 |
| 32 | Le Longeron (Manis) | 47.018959 / -1.04650 | hedge | crop field | 2007 | 14 | 3 | 12 |
| 33 | Le Longeron (Grange) | 47.014237 / -1.02699 | hedge | crop field | 2007 | 16 | 3 | 12 |
| 34 | Montravers | 46.835048 / -0.72040 | hedge | crop field | 2010 | 30 | 6 | 9 |
| 35 | Courlay | 46.776652 / -0.53782 | hedge | crop field | 2010 | 14 | 3 | 9 |
| 36 | Noirlieu | 46.919363 / -0.42997 | hedge | crop field | 2010 | 15 | 3 | 9 |
Fig. 1 - Location map of the elm clone experimental plantations. Tests codes (e.g., “#31”) are formed using Region number (e.g., “#3” for Pays de Loire) followed by the test number in that region (e.g., “#1”).
As indicated in Tab. 2, the previous land use varied from crop fields in most hedge-type plantations to pastures or woodlands in the plot-type plantations. In the crop fields, soil and soil preparation were much better than for forest plantings.
The three plot type tests planted in 2007 and 2008 comprised at least ten clones (totaling 100 plants) of native field elms or U. × hollandica Mill. The hedge type tests contained only 3 to 7 clones, i.e., 15 to 36 elms. All tests were planted on different sites except at Banneville, where the two hedges were planted on the long sides of a hayfield containing the plot.
In their first years, the experimental plantations were measured during winter, generally at the end of their third and fifth year or more frequently. After reaching the stage to become attractive to bark beetles, they were examined in late July or August to monitor DED symptoms. The current height (i.e., height of the highest living leaf) of all trees was measured in this period. The wilt percentage of diseased trees was estimated. From the situation of the symptoms in a part of the tree crown and by comparison with neighbouring trees, the cause of wilting could clearly be attributed to O. novo-ulmi and not to water stress. In doubtful cases with symptoms restricted to a single branchlet, early stage wilt was scored 1% but combined with 0% scores for data analysis because these trees may still be perfectly “healthy” if symptoms had a cause other than DED (e.g., a twig-mining larva). In case of a new infection causing dieback, the height of the tree before infection was measured. From 2016, a “loss of crown size” percentage was also estimated to quantify the dieback of the branch architecture of the tree. This new percentage was needed because some trees have perfectly green leaves (0% wilt) but those rare leaves are restricted to the last living branch of a tree skeleton (95% loss of crown).
With the exception of test #12, the plantations located at Nogent/Vernisson, Central France (Fig. 1) were surveyed nearly every year, and so was the large plot-test #31 near Nantes (Western France), planted and measured jointly with ONF-PNRGF. Apart from the slow-growing hedge #24, the other plantations were generally surveyed by IRSTEA every two years.
Data and statistical analyses were processed with R version 3.5.1 and graphics with Microsoft Excel® (Mac version 14-7-3). Analyses of variance tables are given in Supplementary material (Tab. S1). The large set of data considered at species level (i.e., Ulmus minor of the French national collection) enabled the production of descriptive statistics on DED infection and subsequent effects such as the mortality rate of symptomatic trees. DED infection was studied both in its dynamics since planting (“DED incidence”) and as the resulting situation considered at the time of the plantations’ last assessment (“DED prevalence”).
Except in plot-test #31, fine comparisons between many clones were not possible because of the low number of trees per clone per test and differences in clonal composition among plantations. However, large numbers of trees were available for the two clones planted together in eight experimental plantations and for the 11 clones common to plot-tests #21 and #31. In addition, the frequently planted clones F032mg and F083 could be used as benchmarks to characterize clones with remarkably good or bad results in a test. Symptoms development (i.e., signs of recovery vs. persistence of symptoms or death) was studied on a subset of 122 trees infected at least three years before the last measurement.
Height at five years after planting is presented in order to compare the vigour of trees between the sites. Height in the four-year-old test #31 is also given to enable comparison between clones. No other data on height increment of healthy plants are shown here because growth considered alone is far less important than DED infection, mortality and dieback.
Results
Height variation between eight plantations before infection by DED
Average tree height of clone F032mg was compared five years after planting in eight plantations (Tab. S1.1 in Supplementary material), i.e., before infection by DED except for two cases in test #34. The average height of F032mg exceeded 6 m in the best growth conditions (#35, #23 and #36). It was significantly higher (P<0.05 - Tab. S1.2) on crop field soils (6 hedge-tests; 4.98 m) than in the hayfield and the woodland site suffering high competition with woody plants regrowth (respectively, 3.74 m and 2.71 m).
Height variation between clones before infection by DED in plot-test #31
Average tree height of 20 clones with at least 5 trees was compared four years after planting in plot-test #31 (Tab. S1.3 in Supplementary material), i.e., before infection by DED except for three infection cases. Clones F032mg (3.8 m), F034 (3.1 m), F140 (3.09 m) and F050 (2.98 m) were found significantly taller than F044, F203, F355 and F020, which were under 1.9 m.
Incidence of DED
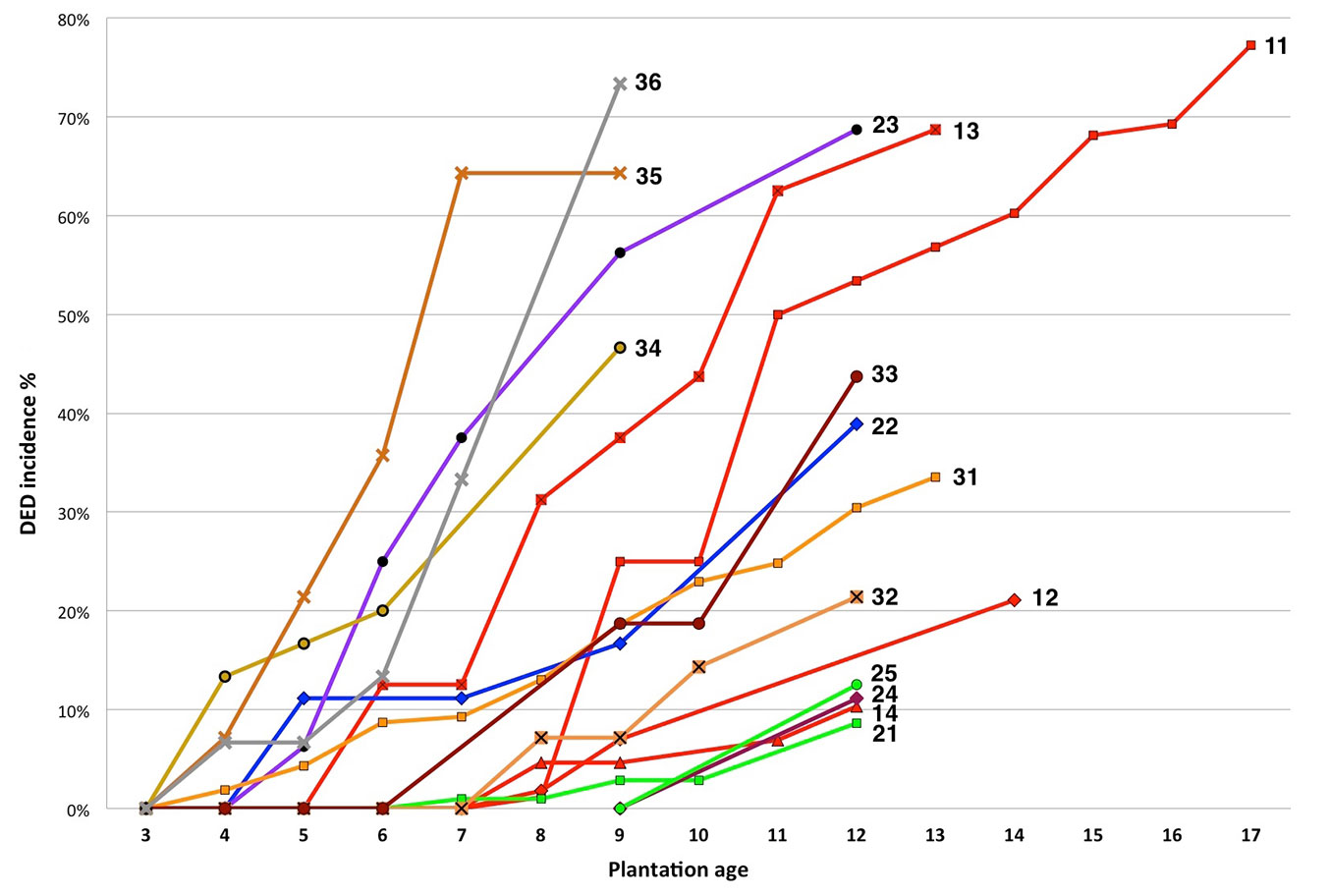
The development of DED infection over time in the 15 plantations where DED was found is shown in Fig. 2. The curves of fast-growing hedge-tests #23, #34, #35 and #36 rise steeply four or five years after planting whereas the curves of large plot-tests #31, #14, #21 and #12 are more gentle and/or rise approximately eight years after plantation. After reaching about 55% incidence, curves tend to shift less steeply.
Fig. 2 - Dynamics of DED incidence in the 15 experimental plantations with infected trees. Each of the 15 curves is terminated by the code of the plantation. DED incidence % is the number of diseased trees over total number of trees.
As regards clones, the final infection rates of 20 clones with at least 5 trees in plot-test #31 were examined in relation with tree heights four years after planting: (i) there was a significant 22% correlation (p = 0.006) between height at age 4 and DED prevalence at last assessment; (ii) altogether, the mean height of the 53 future diseased trees was significantly 15% higher (p<0.05) than the mean height of the 104 trees which remained asymptomatic, but six clones did not follow this trend (Tab. S1.4, Tab. S2.4 in Supplementary material); (iii) significantly taller clones (F032mg, F034 and F140) were frequently infected (≥ 50%) except F050 (33%), whereas two (F044 and F355) of the four significantly smaller clones remained asymptomatic; (iv) two clones were exceptions to the above trend: F056 with a very high infection rate (89%) in spite of a medium mean height (2.37 m) and F028 with no infection in spite of a similar medium mean height (2.43 m).
Another remarkable case of high DED incidence was observed in tests #12, #13 and #14, where clone F250 was infected fewer years after planting and/or more frequently than the other clones in the same tests (data not shown).
DED prevalence variation between tests
At the time of their last assessment, the average prevalence of DED in the experimental plantations was 32.7% (232 diseased trees out of 710), with strong variations between tests (Tab. 3). Average DED prevalence of clone F032mg was compared in eight plantations (Tab. S1.5 in Supplementary material) and ranged from 100% in hedge-test #35 in excellent growth conditions to 11% in plot-test #21 in a hayfield.
Tab. 3 - DED prevalence and mortality of diseased trees per plantation. (Ntot): number of trees; (Ndis): number of diseased trees; (Ndead): number of dead trees; (DED%): DED prevalence (%); (M%): mortality (%) of diseased trees. (‡): at least 9 trees diseased.
| Test code | Age | All Clones | Clone F032mg | |||||
|---|---|---|---|---|---|---|---|---|
| Ntot | Ndis | Ndead | DED% | M% | DED% | M% | ||
| 11 | 17 | 88 | 68 | 20 | 77 | 29 ‡ | 64 | 29 |
| 12 | 14 | 57 | 12 | 3 | 21 | 25 ‡ | 14 | 0 |
| 13 | 13 | 16 | 11 | 3 | 69 | 27 ‡ | 75 | 0 |
| 14 | 12 | 87 | 9 | 3 | 10 | 33 ‡ | - | - |
| 21 | 12 | 104 | 9 | 0 | 9 | 0 ‡ | 11 | 0 |
| 22 | 12 | 18 | 7 | 1 | 39 | 14 | 50 | 33 |
| 23 | 12 | 16 | 11 | 7 | 69 | 64 ‡ | 80 | 100 |
| 24 | 12 | 36 | 4 | 0 | 11 | 0 | 17 | 0 |
| 25 | 12 | 24 | 3 | 0 | 13 | 0 | - | - |
| 26 | 12 | 14 | 0 | 0 | 0 | - | - | - |
| 31 | 13 | 161 | 54 | 1 | 34 | 2 ‡ | 50 | 0 |
| 32 | 12 | 14 | 3 | 1 | 21 | 33 | - | - |
| 33 | 12 | 16 | 7 | 0 | 44 | 0 | - | - |
| 34 | 9 | 30 | 14 | 2 | 47 | 14 ‡ | 80 | 25 |
| 35 | 9 | 14 | 9 | 4 | 64 | 44 ‡ | 100 | 50 |
| 36 | 9 | 15 | 11 | 3 | 73 | 27 | 80 | 50 |
| Total | - | 710 | 232 | 48 | 32.7 | 20.7 | 43 | 32 |
DED prevalence variation between clones
Four views of increasing width in terms of clone numbers and decreasing value in terms of number of trees per clone are presented here: (i) clone F083 was significantly (p<0.05) less frequently diseased (23%) than F032mg (42%) in the eight tests where they are planted together (about 60 trees per clone - Tab. S1.6 in Supplementary material); (ii) clones F056 and F140 were significantly (p<0.05) more frequently diseased (56% and 50%) than F028, F044 and F083 (respectively 0%, 0% and 5%) in plot-tests #21 and #31 where they are planted together (14 to 20 trees per clone - Tab. S1.7); the nine other clones in the same tests did not differ from them, except for F020 (12%) which differed significantly from F056; the average DED prevalence of F032mg was 32%; (iii) clone F203 was very frequently diseased (75%) in contrast with F028 (0%) in plot-test #31 (Tab. S1.8); (iv) in Tab. S2.1 summarizing the DED prevalence of all 38 clones in all plantations, one can take note of the 100% infection rates of clones F040 and F178, planted only in test #11. The DED prevalence means of F032mg and F083 in the same experimental plantation #11 were respectively 64% and 67%.
Symptoms development: wilt, mortality rates and recovery signs
Wilt
The average wilt percentage of the 232 diseased trees (35 clones) in their infection year was 37% (Tab. S2.1 in Supplementary material), ranging from more than 70% for severely defoliated clones to less than 15% for the least defoliated, with even greater variation (standard deviation 29%) between trees of the same clone, e.g., 5% to 95% in the case of clone F034.
Mortality
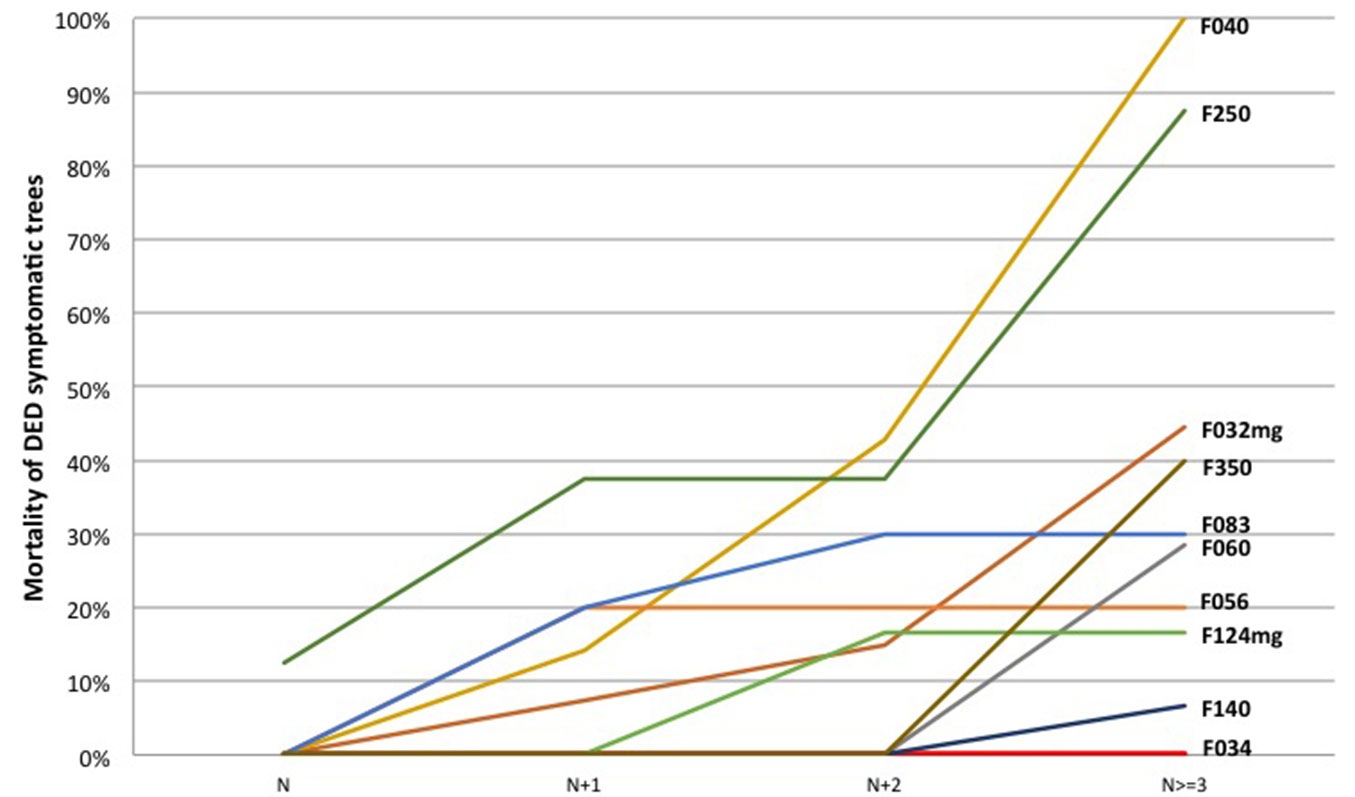
Out of the 122 trees infected at least three years before last measurement, 44 (36%) died in the monitoring period, rarely in the infection year and mostly (25 trees) three years or more after infection (Tab. S2.2 in Supplementary material). A closer focus on the development of mortality in ten clones with at least five diseased trees is shown on Fig. 3. Mortality began in the infection year for clone F250 and was also rapid for F040, reaching 40% in the second year after infection. On the contrary, it had still not begun for F034 in the third year and remained under 10% for F140. Mortality tended also to be delayed for clones F060 and F350, and to remain under 35% for F124mg, F056 and F083.
Fig. 3 - Development of mortality through time in 10 clones with at least 5 diseased trees over post infection years. Clone codes are displayed at the end of each curve. (N): infection year.
Recovery signs
Trees with perfectly green foliage three years after infection can be considered “with recovery signs”. Trees of clone F034 were found with recovery signs in several tests. Fig. 4 shows pictures of a tree of clone F034 in hedge-test #33 on the infection year (80% wilt in 2015), one year later (80% wilt again in 2016) and three years later (no recurrence of wilting in 2018). Even if a few dead shoots are still visible, the July 2018 photograph shows that the tree has fully recovered its ante-infection size.
Fig. 4 - Tree #8 of F034 in hedge-test #33 in the infection year (a), one year later (b) and three years after infection (c). 80% wilt on 30/07/2015; 80% wilt on 16/09/2016 ; 0% wilt on 31/07/2018.
Contrasted trends were observed in three neighbouring trees of F034 in test #11. All of them had fully recovered from initial infection in 2010, when their height ranged between 3.9 m and 4.8 m. A new outburst of wilt appeared on one of them in 2015 and on the two others in 2016, when their height ranged between 7.9 and 8.2 m. At the time of last measurement, only one tree was totally healthy, whereas the two others were very severely wilted (>85%). The same situation was observed with trees of F060 which had fully recovered from the 2010-2013 infections and were hit by new outbursts of wilt after 2015. A few trees of clones F083, F124, F140 and F077 were still showing recovery signs in test #11.
Average mortality of symptomatic trees at the time of plantations last assessment
At the time of their last assessment, the mortality of symptomatic trees in all plantations was 21% (48 dead trees out of 232 diseased trees - Tab. 3). Focusing on the ten tests with at least nine diseased trees, the mortality of symptomatic trees was highest (64% and 44%, respectively) seven years after first DED symptoms in hedge-tests #23 and #35 in the best growth conditions, whereas it was still 0% five years after first symptoms in slow growing plot-test #21. It was 29% in the 17-year-old test #11.
Discussion
DED infection
The reasons for the variation of DED incidence observed between tests are not clear and probably multifactorial. One can assume that environmental conditions favouring tree growth account for the time before trees are high enough to attract massively the vectors of DED (Scolytus spp.), i.e., about 4 years for fast-growing trees and about 8 for slow-growing ones. In addition, the pressure level of the local population of vectors is known to be crucial ([29]) and may play a role in the stronger incidence of DED in small multispecies hedge-tests compared to large monospecies plot-tests, e.g., if bark beetle reservoirs are sufficient to inoculate a large number of the 15 to 36 elms in a hedge-test, but insufficient to inoculate a large portion of the 100 and more elms (161 for test #31) of a plot-test. The hypothesis of infection by mechanical trimming can be rejected because no trimming was carried out in the most diseased hedge #23.
The variation of DED incidence among clones is probably also caused by clonal differences in growth vigour (e.g., the high DED incidence of fast growing clone F032mg in test #31) but not solely, as observed with the contrasted cases of F056 (medium size; high DED prevalence) and F028 (medium size; low DED prevalence). Severely and quickly infected clones F140, F056, F203, F250 and F350 on one hand, and less severely infected F028 and F083 on the other hand, would be good candidates for experiments on clone attractiveness for DED vectors ([21], [2]). Interestingly, F250 was also reported by Girard ([11]) as DED symptomatic in a 4-year-old test.
DED prevalence was very high (77%) in the oldest experimental plantation (17 years) and also in younger but faster growing hedge tests (e.g., over 63% in nine-year-old tests #35 and #36). In addition, clones F040 and F178 had already reached 100% prevalence in the oldest plantation. These facts suggest that DED incidence may increase continuously with time and that all trees of field elm clones will finally be infected by DED. However, the wide range of variation observed between tests and clones gives hope that, under certain site conditions, trees of some clones may escape the disease for several decades. This trend could be observed for clone F083, with one plant out of three still healthy even in tests where DED was frequent. However, no firm result regarding such tolerance will be available before plot-tests #14, #21 and #31 are heavily infected. We recommend that a lighter monitoring of disease infection and tree mortality should be continued in those three tests.
Recovery signs
Clone F034 was able to recover more readily and durably than the other clones and could serve for experiments on recovery ability. Clones F140, F060, F350, F124mg, F056, and F083 also tended to suffer less wilting recurrence over the 3 post-inoculation years. However, such remissions were often brutally stopped by a new outburst of wilt, supposedly a new infection by bark beetles. Investigations are needed to clarify this point because disease recurrence cannot be totally excluded, nor infection by root grafts in the case of plots with 5 m between elm trees. Altogether, the rarity and fragility of remissions give little chance for complete and long-lasting returns to health. Trees of F140 were found able to survive many years after infection but the trees often offered a pathetic appearance as a consequence of past severe crown dieback.
Mortality
Clones F250 and F040 tended to die quickly after infection and could be used, in contrast with clone F034, for research on factors of tolerance to O. novo-ulmi.
The 21% average mortality recorded at last assessments does not give a full picture of the situation because only about 70% of the diseased trees die in less than three years after infection. The 36% mortality rate of the 122 trees infected at least three years before last measurement gives a better indication of the magnitude of the problem, which was totally unknown in artificial inoculations tests, where mortality is rare and restricted to very susceptible clones ([22]). The high mortality (64%) recorded seven years after first DED symptoms in hedge-test #23 is alarming, but may have been exacerbated by the high quality of the soil (previously used for horticulture) and the proximity of a brook, causing the very fast growth of the trees, particularly of clone F032mg. As experimented by Solla & Gil ([25]) and reviewed by Martin et al. ([17], [19]), environmental conditions, particularly water availability, may influence the fate of diseased trees. Anatomical studies comparing wood samples of F032mg from different tests would be needed ([3], [15], [18]) to judge whether the high mortality of clone F032mg in test #23 (four diseased trees, all dead) could be an effect of the very fast growth of the trees in that plantation.
Because the three large plot-tests planted in 2007 and 2008 will continue to produce rare and valuable data on tree long-term survival, the data files and maps and the microsatellite fingerprint of nine clones are supplied as Supplementary material (Appendix 3, Appendix 4), such that new researchers can go back to the field and re-assess these experimental plantations in the future. High definition aerial or satellite pictures may be helpful to trace back wilt and mortality.
Practical implications of results for the conservation of genetic resources
The field testing activity reported here is part of the French elm conservation programme, not only in the scope of gaining knowledge on the variability of DED susceptibility in native field elms but also for evaluating strategies for ex-situ conservation as well as for dynamic conservation. Because the management of clonal banks is costly in terms of land occupation and annual trimming, it is not always possible to maintain all clones as growing material in a nursery. Reducing clone number to the size of a core-collection is a solution frequently adopted, possibly in combination with cryopreservation ([8]). Another alternative, or at least an interesting supplementary option, might be a network of plantations of regional clones. The case of plot-test #21 offers the example of a 120-elm plantation assembling 12 clones, mostly from the local region (10 Normandy clones and two from Poitou-Charentes); the land belongs to the Departmental Council of Calvados and the regional environmental NGO (Crepan) ensures the monitoring of the test. Twelve years after plantation, no clone has been lost and the test can be used for education purposes and possibly for seed collection. This good situation, however, may be challenged by DED in the future because prevalence may increase dramatically with the vectors’ population dynamics ([14]). If many clones are lost and/or most of the trees offer a severely diseased appearance, the conservatory role of the plantation may come to an end. In this hypothesis, a field conservation plantation should rather be considered as a supplementary conservation method and not an alternative to nursery clonal banks maintained unattractive to DED vectors as low hedges. However, if conservation plantations are planted as randomized single tree plots and do not suffer too many losses, they can be used for studies on clones and also supply seed lots for hedge reconstruction. The genetic diversity of the seed lots would benefit from the plot arrangement facilitating panmixia and from the large genetic diversity assembled in the plantation. A minimum number of 30 clones with frequent recovery in inoculation tests could be recommended, and this would probably ensure a larger diversity than natural stands shaped by clonality because of the high capacity of U. minor for vegetative reproduction by root-suckers ([9], [6]). Soil and other environmental conditions should be fair but not excellent in order to avoid an excessively fast growth of the trees which would increase their susceptibility to DED, as reviewed by Martin et al. ([19]) and suspected for hedge-test #23. A flat hayfield, as for plot-test #21, would provide the proper soil and environmental conditions, and make the access to trees easy for scientific monitoring and for seed collection for hedge reconstruction projects. In contrast, a nursery clonal bank does not allow trees to grow freely, and does not facilitate seed production nor panmixia.
Genetic resources conservation should not be restricted to the static preservation of genotypes adapted to the present or past situation ([10]). On the contrary, dynamic conservation should stimulate the process of adapting tree populations to changes in their environment. Dynamic conservation is usually carried out through natural regeneration in natural populations but a kind of “dynamic restoration” could be applied in the case of U. minor in field hedgerows ([7]). Collin et al. ([8]) do not recommend the planting of allochthonous material in riparian field elm populations, supposedly natural and large enough to maintain themselves with profuse suckering and young mature trees. In contrast, they suggest that planting genetically diverse material (clones and/or seedlings) could help reinforce the gene pool of hedges severely depopulated by DED. Such inputs, provided they remain quantitatively small and genetically diverse, would not be a truly new practice for field hedgerows, which have been largely shaped by humans for centuries ([6]). In Spain, Martin et al. ([17]) advocated the use of the seven native tolerant field elm clones registered in the country ([16]).
The hedge-tests presented in this study may be considered as elm dynamic restoration experiments because the material is native, genetically and phenotypically diverse and likely to live long enough to produce flowers (Collin et al., in preparation) and exchange pollen with other elms in the plantation and also in their neighbourhood, hence contributing seedlings (i.e., new genotypes) that will be subjected to natural selection and provide fuel for on-going adaptation process. However, the results presented here show that the French native material is far from being as tolerant as the Spanish registered clones. Consequently, there is a risk of loss of several clones, causing a decrease in the genetic diversity of the plantation. To remedy this risk, one should increase the number of clones and add seedlings of local origin, even if this addition results in diminishing the number of trees per clone. This increase would be facilitated by local plant certification initiatives promoting the use of seed batches collected in at least three sites in the same region ([28]). Moreover, the question of introducing material from a distant region has to be addressed in the context of climate change because assisted migration ([1]) may facilitate adaptation of trees to new environmental conditions, including new pests and diseases.
Practical implications of results for hedge reconstruction
The high incidence of DED and high mortality observed in several experimental plantations suggest that hedge reconstruction projects with French U. minor material cannot be driven by technical and economical interests only, but rather by more intimate cultural reasons relating to heritage value and landscape aesthetics. For instance, the most successful U. minor clones could be suitable for planters interested in restoring mature elms for biodiversity (e.g., for the rare Satyrium w. album butterfly) but refusing Eurasian DED resistant cultivars because of introgression risks towards native elm genetic resources. This issue is discussed in Collin et al. (in preparation), which also examines a combination of height and health criteria as an indicator of the plantation success seen from a planter’s point of view.
Conclusion
The results presented here bring novel information on DED incidence in experimental plantations of a broad array of U. minor clones of the French national elm collection. Variation in DED incidence was observed between clones and according to environment. Differences in symptoms development were detected, highlighting the better recovery ability and longer survival of a few clones (Tab. 4).
Tab. 4 - Overview table of the performance of 14 clones in experimental plantations and inoculation tests. (GV): growth vigour in plantations; (DED%): lower or slower incidence of DED in plantations; (RS): recovery signs and/or longer survival of diseased trees in plantations; (IT): frequent recovery or limited symptoms in artificial inoculation test as coded in column IT of Tab. 1. Superior “+” or negative “-” performance signs are followed by sign “*” when significantly different. Signs between brackets are used for field observations not supported statistically. (0): non-significant differences.
| Clone | GV | DED% | RS | IT |
|---|---|---|---|---|
| F028 | 0 | +* | (+) | (+) |
| F032mg | +* | -* | 0 | (+) |
| F034 | +* | (-) | (+) | (+) |
| F040 | 0 | 0 | (-) | (-) |
| F056 | 0 | -* | (+) | (+) |
| F060 | 0 | 0 | (+) | (+) |
| F077 | 0 | 0 | (+) | (+) |
| F083 | 0 | +* | (+) | (+) |
| F124mg | 0 | 0 | (+) | (+) |
| F140 | +* | -* | (+) | (-) |
| F178 | 0 | 0 | (-) | ? |
| F203 | -* | (-) | 0 | ? |
| F250 | 0 | (-) | (-) | (-) |
| F350 | 0 | 0 | (+) | (+) |
These results provide foundations for new research and breeding activities using native European clones with contrasted levels of infection rate or recovery ability. As suggested by Martin et al. ([19]), combining low attractiveness to bark beetle feeding with resistance to O. novo-ulmi might considerably enhance the field performance of elm cultivars.
Practical conclusions for genetic resources conservation consist in recommendations for the establishment of conservation plantations of regional clones: fair but not excellent site conditions in order to avoid an excessively fast growth of the trees, preferably a flat hayfield making access to trees easy, a minimum number of 30 clones, a single tree plot design facilitating panmixia and the collection of reliable scientific data.
The high prevalence of DED and the high mortality observed in several experimental plantations suggest that hedge restoration projects with native French elms must be motivated by heritage value rather than by technical or economic interest.
Acknowledgments
The elm clone testing programme of IRSTEA has received the financial support of the French Ministry of Agriculture; the plantings of tests #21 and 31 were respectively carried out in collaboration with the Departmental Council of Calvados and J. Rieffel Agriculture School (F44800 Saint-Herblain).
The cooperation of Monique Guibert and Aurore Desgroux is acknowledged in arranging T. Pozzi’s assistance with data analysis after E. Collin’s retirement.
The authors thank Luc Harvengt and Aurore Desgroux for their improvements to the manuscript, and Andrew Brookes, Timothy and Peter Stafford for their kind linguistic assistance. Deep gratitude is expressed to the three anonymous reviewers for their very helpful comments and suggestions, and to the diverse people, too many to mention by name, who have assisted with the testing presented in this work.
This paper is dedicated to Hans Heybroek (1927-2022), who spent his long career studying and breeding elms. He was universally appreciated for his immense knowledge and unstinting kindness.
Authors’ contributions
EC instigated the elm clone field testing programme of IRSTEA, participated in field work, processed the data with Excel, and wrote the manuscript; TP processed the data with R and assisted EC in drafting the manuscript; CJoyeau and SM carried out the field work; MR propagated the plant material and assisted field work; CJoly directed the joint elm clone field testing programme of CREPAN and IRSTEA, and participated in field work with the IRSTEA team.
List of abbreviations
CEMAGREF: Centre d’Etudes du Machinisme Agricole, du Génie Rural, des Eaux et Forêts; INRA: Institut national de la recherche agronomique; INRAE: Institut national de recherche pour l’agriculture, l’alimentation et l’environnement; IRSTEA: Institut national de recherche en sciences et technologies pour l’environnement et l’agriculture; ISSRs: Inter-simple sequence repeat molecular markers; RAPDs: Random Amplified Polymorphic DNA molecular markers.
References
Gscholar
Gscholar
Gscholar
Gscholar
Gscholar
Supplementary Material
Authors’ Info
Authors’ Affiliation
Tiffani Pozzi 0000-0003-2540-3757
Cécile Joyeau
Stéphane Matz
INRAE, UR EFNO, F-45290 Nogent-sur-Vernisson (France)
Office National des Forêts, Pôle National des Ressources Génétiques Forestières, F-44290 Guémené-Penfao (France)
CREPAN, 8 rue Germaine Tillion, F-14000 Caen (France)
Corresponding author
Paper Info
Citation
Collin E, Pozzi T, Joyeau C, Matz S, Rondouin M, Joly C (2022). Monitoring of the incidence of Dutch Elm Disease and mortality in experimental plantations of French Ulmus minor clones. iForest 15: 289-298. - doi: 10.3832/ifor3820-015
Academic Editor
Alberto Santini
Paper history
Received: Mar 21, 2021
Accepted: Jun 13, 2022
First online: Jul 29, 2022
Publication Date: Aug 31, 2022
Publication Time: 1.53 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2022
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 28636
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 24636
Abstract Page Views: 2037
PDF Downloads: 1506
Citation/Reference Downloads: 4
XML Downloads: 453
Web Metrics
Days since publication: 1288
Overall contacts: 28636
Avg. contacts per week: 155.63
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2022): 2
Average cites per year: 0.50
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Comparison of commercial elm cultivars and promising unreleased Dutch clones for resistance to Ophiostoma novo-ulmi
vol. 8, pp. 158-164 (online: 07 August 2014)
Research Articles
Genetic variation and heritability estimates of Ulmus minor and Ulmus pumila hybrids for budburst, growth and tolerance to Ophiostoma novo-ulmi
vol. 8, pp. 422-430 (online: 15 December 2014)
Research Articles
Seven Ulmus minor clones tolerant to Ophiostoma novo-ulmi registered as forest reproductive material in Spain
vol. 8, pp. 172-180 (online: 13 August 2014)
Review Papers
Genomics of the Dutch elm disease pathosystem: are we there yet?
vol. 8, pp. 149-157 (online: 07 August 2014)
Review Papers
Dutch elm disease and elm bark beetles: a century of association
vol. 8, pp. 126-134 (online: 07 August 2014)
Review Papers
Avoidance by early flushing: a new perspective on Dutch elm disease research
vol. 2, pp. 143-153 (online: 30 July 2009)
Technical Reports
Conservation and use of elm genetic resources in France: results and perspectives
vol. 13, pp. 41-47 (online: 03 February 2020)
Review Papers
Implementing the dynamic conservation of elm genetic resources in Europe: case studies and perspectives
vol. 8, pp. 143-148 (online: 07 August 2014)
Research Articles
Genetic variation of Fraxinus excelsior half-sib families in response to ash dieback disease following simulated spring frost and summer drought treatments
vol. 9, pp. 12-22 (online: 08 September 2015)
Review Papers
Ulmus laevis in the Iberian Peninsula: a review of its ecology and conservation
vol. 8, pp. 135-142 (online: 07 August 2014)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword