Coping with spring frost-effects on polyamine metabolism of Scots pine seedlings
iForest - Biogeosciences and Forestry, Volume 10, Issue 1, Pages 227-236 (2017)
doi: https://doi.org/10.3832/ifor2003-009
Published: Jan 27, 2017 - Copyright © 2017 SISEF
Research Articles
Abstract
Polyamines (PA) are ubiquitous polycations known to be involved in several phases of plant development as well as in tolerance to abiotic stresses. Phenols are complex secondary metabolites produced via the phenylpropanoid pathway that contain, e.g., cell wall compounds and antioxidants. Phenols are known to enhance chilling tolerance of plants. PA and phenolic pathways are connected via conjugation. In boreal coniferous forests spring frost has been considered to have severe effects on the survival of tree seedlings. Such effects are likely to increase in the future. The present study focuses on the role of PA and phenylpropanoid syntheses in the coping strategies of Scots pine exposed to cold temperatures during the vulnerable early seedling phase in late spring and early summer. We found that spring frost affects the expression of genes regulating PA metabolism and phenylpropanoid synthesis differently in above and below ground parts of the seedlings, whereas PA or phenol contents in tissues were not affected. The results suggest that Scots pine seedlings may not have time to develop metabolite level responses during a short period of freezing stress and, therefore, the originally different PA levels, especially in roots, may influence the tolerance of Scots pine seedlings to spring frost.
Keywords
Introduction
Scots pine (Pinus sylvestris L.) is one of the most widespread coniferous species in boreal zones and important to Fennoscandian forestry. Due to the first Finnish forest improvement act in 1928 large mire areas were drained for the forestry during the ’30s. Therefore, large drained peatland areas in Finland are approaching the age of renewal. Peat is a challenging growth substrate having its own special characteristics, such as hydrological properties, temperature conditions and poor nutrient content, which has to be considered upon forest regeneration ([38]). Investigation of freezing stress induced transcription and metabolite level changes in pine seedlings enhance understanding of stress responses during the vulnerable early growth phase.
Paradoxically, a warming climate has been predicted to increase frost damages to trees in boreal forests. Due to climate change higher winter and spring temperatures will promote earlier dehardening and growth onset, thus extending the risk period of frost damage ([3], [16]). For example, needles of Scots pine from previous year retain some of their frost hardiness after the growth of apical meristems has begun ([27]). Tree organs are suggested to be more susceptible to frost damage during the early phenological development than organs from the previous year ([28]). Therefore, during the post-germination growth first year Scots pine seedlings are particularly vulnerable to frost and tolerance is governed by both its genetic potential as well as by environmental factors ([19]). The origin of the plant material has an impact on the risk of frost damage in following regeneration ([26]).
In Finland, nights at the beginning of June are characterized by clear skies, calm weather and low ambient temperatures, which lead to plants growing near the ground frequently suffering from below zero temperatures. Rikala & Repo ([36]) observed that 8 hours below -5 °C temperature was lethal to 50% of the first year Scots pine seedlings. Moreover, growth of Scots pine roots was inhibited if the soil temperature around roots declines below +5 °C ([37]).
Plant responses to cold have been extensively studied at transcriptional and metabolite levels ([6], [40]). The primary site to sense freezing temperatures are cell membranes, where a cascade of cellular processes is initiated ([45]). Sugars, amino acids, organic acids, PAs and lipids are all actively regulated in response to stress in plant cells ([24]). Cold temperatures trigger the expression of the CBF (gene family with a special C-repeat binding factor in their domain) family of transcription factors, which in turn activate many downstream genes that facilitates freezing tolerance in plants ([40]). Cold stress activates the inducer of CBF expression 1 protein (ICE1) which leads to a cold acclimation cascade in cells ([6]). However, some proteins of the family R2R3-MYB can have antagonistic effect with ICE1 proteins to cold induced genes ([6]). R2R3-MYB genes are transcription factors and belong to a large gene family ([10]), which, in conifers, can directly target genes coding for key enzymes from the interconnected pathways of amino acids and phenylpropanoids ([4]). For example, R2R3-MYB8 up-regulates the carbohydrate, shikimate and monolignol pathways in white spruce (Picea glauca [Moench] Voss - [4]) and the phenylalanine pathway in maritime pine (Pinus pinaster -[8]).
The phenylpropanoids are a diverse family of organic compounds that are synthesized by plants from the amino acid phenylalanine. The first step in phenylpropanoid synthesis is the conversion of phenylalanine to cinnamic acid by phenylalanine ammonia lyase (PAL). Phenolic compounds produced via the phenylpropanoid pathway are related to defense and / or cell wall structure, like lignin, flavonoids and stilbenes which are known to accumulate under abiotic and biotic stresses also in pine tissues ([5], [15]). The stilbenes, pinosylvin and pinosylvin 3-O-methyl ether (PSM) are phytoalexins in young pine seedlings, produced from cinnamic acid-CoA (see Fig. S1 in Supplementary material) ([7]) and are characterized for their antioxidant activities, especially their free radical scavenging properties ([7]).
PAs, putrescine (Put), spermidine (Spd), spermine (Spm) and thermospermine (Tspm), are small nitrogenous compounds that have a prominent role in cold stress regulation in plants ([1]). PA homeostasis in pines is regulated by biosynthesis, catabolism, back conversion and conjugation (see Fig. S2 in Supplementary material). Chilling damage can in several plant species be prevented by increasing PA levels during the cold response ([1]). PAs also have protective effects on membranes and cellular functions against stress ([1]). Furthermore, transgenic white pine (Pinus strobus L.) expressing the pathogen and freezing tolerance related protein 1 (CaPF 1) of pepper (Capsium annuum) had higher PA levels as compared to wild type, and was tolerant to freezing, salt and dehydration stress ([43]). PAs conjugate with phenols, reducing their polarity and hydrophility ([2]). The phenolamides, also called hydroxycinnamic acid amides (HCAA), are a group of phenolic derivatives formed by conjugation of PAs with hydroxycinnamoyl acids ([12]), thus forming a link between PA and phenolic metabolism (see Fig. S1 in Supplementary material).
S-adenosylmethionine synthetase (SAMS) is the key enzyme catalyzing the formation of S-adenosylmethionine (SAM), which is a precursor of PAs and ethylene. Transfer of the methyl group of SAM to hydroxyl group of diverse phenolic compounds is important for plant defense and signaling ([9], [14]). For example, PSM is a methylated product of pinosylvin. SAMS as well as pinosylvin synthase (psSTS) have been shown to be up-regulated during cold acclimation of Scots pine ([20], [25]). In plants, dehydration and low temperatures induce the expression of dehydrins which belong to the group II of late embryogenesis abundant proteins (LEA family - [50]). Dehydrins stabilize membranes ([22]) and the accumulation of dehydrins in tissues in response to dehydrative stress enhances survival of Scots pine at low temperatures ([23]). Expression of catalase (CAT) is related to oxidative stress in Scots pine tissues ([48], [31]).
The aim of the present study was to evaluate the role of PA and phenylpropanoid metabolism in spring frost coping strategies of Scots pine seedlings. Free and soluble conjugated PAs and the expression of the PA synthesizing genes arginine decarboxylase (ADC), spermidine synthase (SPDS), thermospermine synthase (ACL5), S-adenosylmethionine decarboxylase (SAMDC) as well as PA catabolizing genes diamine oxidase (DAO) and polyamine oxidase (PAO) were measured from the needles and roots. The expression of ICE1, SAMS, LEA and CAT genes were analyzed to indicate reactions to stress in the seedlings. Effect of freezing on phenylpropanoid and stilbene synthesis was investigated by measuring the expression of the transcription factor gene (R2R3-MYB8), phenylalanine ammonia lyase (PAL) gene and psSTS gene and the total amount of phenolic compounds as well as the content of pinosylvin and PSM.
Moreover, we show here that spring frost affects PA metabolism and phenylpropanoid synthesis differently in above and below ground parts of the Scots pine seedlings. Furthermore, our results suggest that the coping with spring frost includes specific defense mechanisms because the stress responses which enhance plant tolerance during long-lasting freezing stress may not be as useful in the face of sudden and brief spring frost.
Material and methods
Plant material and growth chamber cultivation prior to frost experiment
Scots pine (Pinus sylvestris L.) seeds originated from three controlled crossings made for breeding purposes in southern Finland (Punkaharju, 61° 45′ 20″ N, 29° 23′ 20″ E) with northern (Kolari, 67° 17′ N, 23° 46′ E) mother grafts and pollen clones. The crossings, female graft (P74) × male pollen clone (P107 - G4-92-360), female graft (P79) × male pollen clone (P74 - G4-92-361) and female graft (P206) × male pollen clone (P111 - G4-92-362) formed the full-sib families S1, S2 and S3, respectively. Scots pine seeds were sown on peat substrate (Kekkilä Metsätaimiturve M1L) on comb flowerpots containing six 57 × 24 cm cells. Altogether 42 seeds per crossing, seven seeds per cell, were sown per treatment. Thereafter the seeds were germinated for nine weeks in a growth chamber. For PA, qPCR and phenolic measurements four biological replicates of root and needle tissue were used. The photoperiod in the growth chamber was 6 h darkness and 18 h daylight, light intensity was 180-200 µmol m-2 s-1 and temperature 25 °C. Seedlings were fertilized with nutrients [KH2PO4 25 mg l-1, (NH4) 2HPO4 12.5 mg l-1, CaCl2 2.5 mg l-1, NaCl 1.25 mg g-1, MgSO4 × 7H2O 7.5 mg l-1, FeCl3 × 6H2O 0.6 mg l-1] added to irrigation water. During the growth period, the water content of the peat was maintained at 50-70% as measured by the Theta Probe tension meter (MEA, Magill South, Australia).
Spring frost experiment
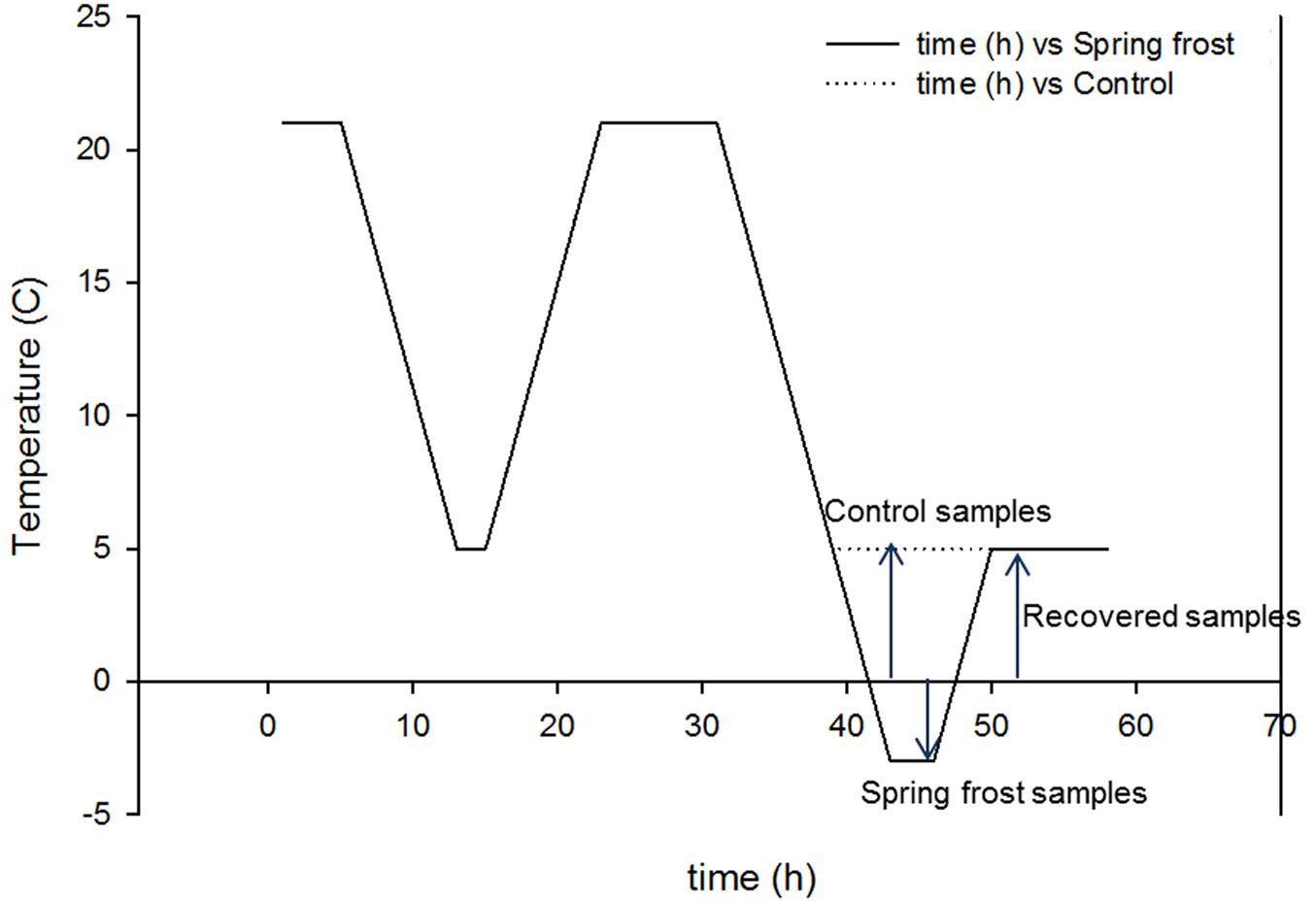
In conifers, the conditions prevailing during sexual reproduction and zygotic embryogenesis appear to adjust the adaptive performance of the progeny by the mechanism that may regulate cold acclimation ([49]). Therefore, the spring frost conditions in the present study were based on the climate data from Punkaharju during the spring or early summer. At the spring time, before pine seeds start to germinate, the temperature stays continuously above 10 °C degrees ([32]). Thereafter the temperature typically stays above zero degrees and freezing temperatures potentially exist only shortly during the late hours of the night. Thus, nine weeks from sowing the seedlings were subjected to short freezing stress in growth chambers (Conviron CMP 3246, Saskatoon, Canada; light intensity in chambers was ca. 150 µmol m-2 s-1 and humidity was 80%). Before the experiment, a one-day acclimation period was used (Fig. 1). For the first night, temperature was decreased from +20 °C to +5 °C for two hours and subsequently raised to +20 °C for the day time at a rate 2 °C h-1. During the second night, temperature was decreased to +5 °C in the control treatment and to -3 °C in the frost treatment. Seedlings were exposed to -3 °C for two hours before sampling. After freezing, temperature was raised to +5 °C for one hour, after which samples were collected and referred as a recovery state. Sampling involved 100 mg of root and needles for RNA and PA measurements, and 100-200 mg for phenol contents. Samples were immediately frozen in liquid nitrogen.
Polyamine analysis
The PAs were extracted from four needle and four root tissue samples from all treatments and crossings with 5% (w/v) perchloric acid. Crude extracts for free PAs and hydrolyzed supernatant for perchloric acid-soluble conjugated PAs were dansylated and separated by HPLC according to Sarjala & Kaunisto ([39]) and Fornalé et al. ([11]). PA concentrations were expressed as nmol g-1 fresh weight of plant tissue. As thermospermine (Tspm) is an isomer of Spm and the present study does not distinguish between Spm and possible Tspm, they are both included in the same fraction and hereafter referred to as Spm.
Sequencing
The polymerase chain reaction (PCR) primers for the amplification of the cDNA fragments of the putative Scots pine ICE1 and R2R3-MYB8 are presented in Tab. S1 (Supplementary material). The primers of the putative ICE1 gene were designed based on the expressed sequence tags of loblolly pine (Pinus taeda L.) [GenBank: DR384895.1], which was selected on the basis of similarity at the nucleotide level with genes isolated earlier from other plant species. The primers of the Scots pine putative R2R3-MYB8 gene were based on the mRNA sequence of the R2R3-MYB8 gene isolated from P. taeda [GenBank: DQ399057]. Fragments with a length of six to seven hundred base pairs were amplified by standard PCR, using seedlings root cDNA as a template and DNA Polymerase (5U µl-1 - Biotools B&M Labs, S.A. Madrid, Spain). The fragments with appropriate length were gel-purified using the purification kit Wizard SV Gel and PCR Clean-Up (System Promega Corporation, Madison, USA) and sequenced by Macrogen Europe (Meibergdreef 31, 1105 AZ Amsterdam Zuid-oost, Netherlands) with an Applied Biosystems 3730xl DNA Analyzer. To ensure that the correct genes were amplified the partial coding sequences were aligned with the protein sequences of other plant species by using a translated nucleotide query (see the alignments in Fig. S3 and Fig. S4 of the Supplementary material).
RNA isolation and reverse transcription
Total RNA was extracted from 100 mg of Scots pine tissue samples (four biological replicates) using a total RNA purification PureLinkTM Plant RNA Reagent (Invitrogen Corporation, California, USA) according to the manufacturer’s instructions. RNA samples were treated with an rDNase set (Magherey-Nagel, Duren, Germany) in order to eliminate contaminating genomic DNA. The amount of DNase used to produce DNA-free RNA samples was three times higher than recommended by the manufacturer. The RNA samples were purified with the NucleoSpin® RNA Clean-Up kit (Macherey-Nagel, Duren, Germany). RNA integrity was checked by agarose gel electrophoresis where two sharp bands (28S rRNA and 18S rRNA) were seen on the gel. If the sample was not intact, the RNA was re-extracted. RNA yields were measured three times with OD260 analysis using a spectrophotometer (Jenway Genova MK2 Life Science Analyser, Dunmow, Essex, UK). One μg of each RNA sample was used for the gene expression analyses. cDNA was prepared from 1 μg of total RNA, which was reverse transcribed using a SuperScript VILOTM cDNA synthesis kit (Invitrogen Corporation, California, USA).
Quantitative real-time PCR analysis
The PCR primers for the expression studies of the PA biosynthesis genes ADC [GeneBank: HM236823], SPDS [GeneBank: HM236827], SAMDC [GeneBank: HM236826], ACL5 [GeneBank: HM236828], DAO [GeneBank: HM236829] and PAO [GeneBank: HM236830] gene were based on the Scots pine PA gene sequences. Similarly, LEA [GeneBank: FJ201571], CAT [GeneBank: EU513163], PAL [GeneBank: AF353986.1], psSTS [GeneBank: S50350.1] and SAMS [GeneBank: JQ970126] primers were based on the known gene sequences of Scots pine. Sequences investigated in the present study were used to design real-time PCR primers of the ICE1 and R2R3-MYB8 genes. The expression of the PA genes, the stress related genes LEA, SAMS and CAT and the phenol synthesis related genes R2R3-MYB, PAL and psSTS were normalized by the gene expression values derived from two different reference genes, ubiquitin (UBQ) [GeneBank: AF461687] and alfa tubulin (TUBA) [GeneBank: FN546172]. All primers and GeneBank accession numbers are listed in Tab. S1 (Supplementary material).
The quantification of mRNA with real-time PCR was performed in 20 μL of reaction mixture composed of 2 μL of cDNA, Light Cycler® 480 SYBR Green I Master Mix, and 100 nM gene-specific primers. PCR amplification was initiated by incubation at 95 °C for 10 min followed by 40 cycles: 30 s at 95 °C, 1 min at 58 °C, and 1 min at 72 °C. The PCR conditions were optimized for high amplification efficiency (90%) for all primer pairs used. Every PCR reaction was done in triplicate to control for the variability of PCR amplification.
The relative gene expression values of the PA genes, the stress related genes and the phenol synthesis related genes were produced by using two different reference genes, UBQ and TUBA. Target/reference ratios were counted by Advanced Relative Quantification method using the software LightCycler 480 version 1.5.0.39. In some samples the target/reference ratios varied depending on the reference gene. To diminish the effect of the reference gene on the final expression values of the target gene, average of target/reference ratio values produced with UBQ and TUBA were used to evaluate the expression level of the target gene.
Total phenol content and stilbenes
For the analysis of secondary metabolites 100-200 mg of needles or roots (four biological replicates) were ground in 100% methanol (1.5ml/100mg), transferred into a test tube and mixed by vortex. After 15 min of incubation on ice the sample was centrifuged at 10.400 × g (Eppendorf Centrifuge 5804 R) for 10min. The supernatant was filtered with a 0.2 µm filter (Syringe Filter, 25 mm, Nylon Membrane) and divided to 2 ml portions into Eppendorf tubes. The samples were vacuum-dried and centrifuged at +45 °C for ca. 5 h. The pellets were stored at -80 °C.
Total phenol contents were analyzed according to the Folin-Ciocalteu procedure ([41]) modified for use on 96-well microplates (BD Labware, Franklin Lakes, NJ. USA), as described in Viitala et al. ([47]). Gallic acid (Sigma-Aldrich) dilutions of 500, 250, 100 and 25 mg l-1 in deionized water were prepared for standard solutions from a stock solution made by dissolving 500 mg gallic acid in 10 ml of ethanol and diluting with deionized water to 100 ml. From standards and samples 1/50 dilutions were used for the measurements.
For HPLC analysis the samples were dissolved to 500 µl of 50% methanol and filtered with 0.2 µl filter. HPLC analysis was performed using a Shimadzu Prominence Liquid Chromatograph system including LC-20AP pumps, a UFLC Prominence Communication Bus Module CBM-20A, a Prominence Photo diode array SPD-M20A Detector (Shimadzu Manufacturing Inc., USA) and a Waters XBridge C18 5 µm 4.0 × 150 mm column (Waters Corporation, USA). The wavelengths of the detector were set at 270 and 320 nm. The stepwise HPLC mobile phase conditions were as follows: initial methanol content 2% increased to 20% at 10 min, to 30% at 20 min, increased to 45% at 33 min, increased to 95% at 35 min and then held in 95% for 10 min; the injection volume was 25 µl. Pinosylvin (Arbonova) and pinosylvin monomethyl ether (Arbonova) were used as external standards to quantify the concentrations of the compounds in pine tissues.
Statistical analysis
Data were analyzed with linear mixed models in program R with the package “nlme” and function “lme” ([33], [35]). PAs, phenols (pinosylvin, PSM and total phenol contents) as well as gene expressions were entered as response variables. Treatment was entered as explanatory variable, with two levels for PA and phenol contents (1= control, +5 °C; 2= freezing, -3 °C), and three levels for gene expression (1= control, +5 °C; 2= freezing, -3 °C; 3= recovery, to +5 °C). The origin of the seedlings was entered as a random factor (three families, S1, S2 and S3). Data were log-transformed when necessary to meet assumption of normality.
Results
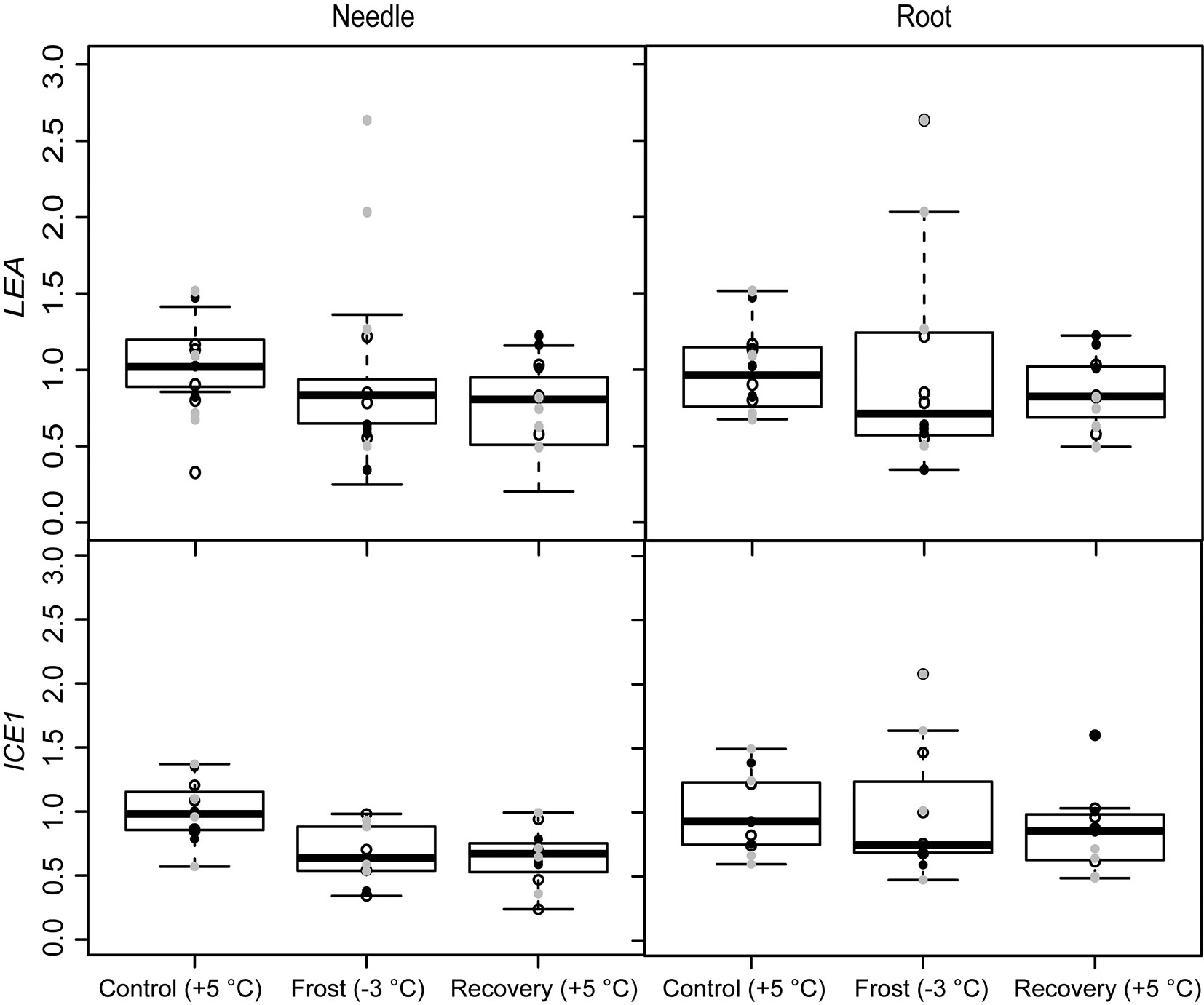
The seedlings did not exhibit pronounced responses to stress induced by spring frost conditions. The only stress related genes affected by the frost treatment in the present study were LEA and ICE1, whose expression decreased in the needles (Fig. 2 and Tab. 1). The expression of ICE1 remained low also after recovery. Expression of the other stress related genes, SAMS and CAT, did not change during the experiment (Tab. 1).
Fig. 2 - Expression of the freezing stress related genes (ICE1) and (LEA), under spring frost (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) in Scots pine needles and roots. Open circles indicate the results from the family S1, black solid circles indicate family S2 and grey circles family S3. Boxplot represents min and max values, first and third quartiles and mean value.
Tab. 1 - The effects of freezing stress treatments (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) on the gene expression results of needles and roots of Scots pine seedlings (linear model). Coefficients with 95% confidence intervals (CI) are presented. Coefficients with CIs not including zero are significant (indicated with “S”). See also Fig. 2 and Fig. 5. (a): difference between control and freezing stress; (b): difference between control and recovery treatment; (*): log transformation before model estimation.
| Needle | Control | Frost a | Recovery b | Root | Control | Frost a | Recovery b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Est | 95% CI | Est | 95% CI | Est | 95% CI | Needle | Est | 95% CI | Est | 95% CI | Est | ||
| CAT | 1.01 | 0.86, 1.14 | -0.17 | -0.36, 0.03 | -0.1 | -0.30, 0.09 | CAT | 1 | 0.88, 1.12 | 0.01 | -0.16, 0.18 | -0.13 | -0.30, 0.04 |
| LEA | 1.00 | 0.84, 1.16 | -0.2 | -0.43, 0.03 | -0.26s | -0.49, -0.03 | LEA* | -0.04 | -0.27, 0.2 | -0.14 | -0.47, 0.19 | -0.16 | -0.49, 0.17 |
| SAMS | 1.00 | 0.72, 1.28 | -0.11 | -0.45, 0.23 | 0.06 | -0.28, 0.4 | SAMS* | -0.04 | -0.19, 0.11 | 0.11 | -0.11, 0.32 | 0.03 | -0.19, 0.24 |
| ICE1 | 1.00 | 0.87, 1.13 | -0.33s | -0.51, -0.15 | -0.36s | -0.53, -0.18 | ICE1* | -0.04 | -0.25, 0.17 | -0.07 | -0.36, 0.23 | -0.18 | -0.47, 0.12 |
| R2R3-MYB8 | - | - | - | - | - | - | R2R3-MYB8* | -0.06 | -0.33, 0.22 | 0.1 | -0.23, 0.44 | 0 | -0.33, 0.33 |
| PAL* | 1 | 0.63, 1.37 | 0.08 | -0.33, 0.48 | 0.3 | -0.11, 0.71 | PAL | 1 | 0.83, 1.17 | 0.24s | 0.02, 0.47 | 0.23s | 0.01, 0.46 |
| psSTS* | -0.24 | -0.71, 0.23 | 0.22 | -0.45, 0.89 | 0.43 | -0.24, 1.10 | psSTS* | -0.09 | -1, 0.83 | 0.01 | -0.75, 0.76 | 0.36 | -0.40, 1.12 |
Effects of spring frost on PA metabolism of the seedlings
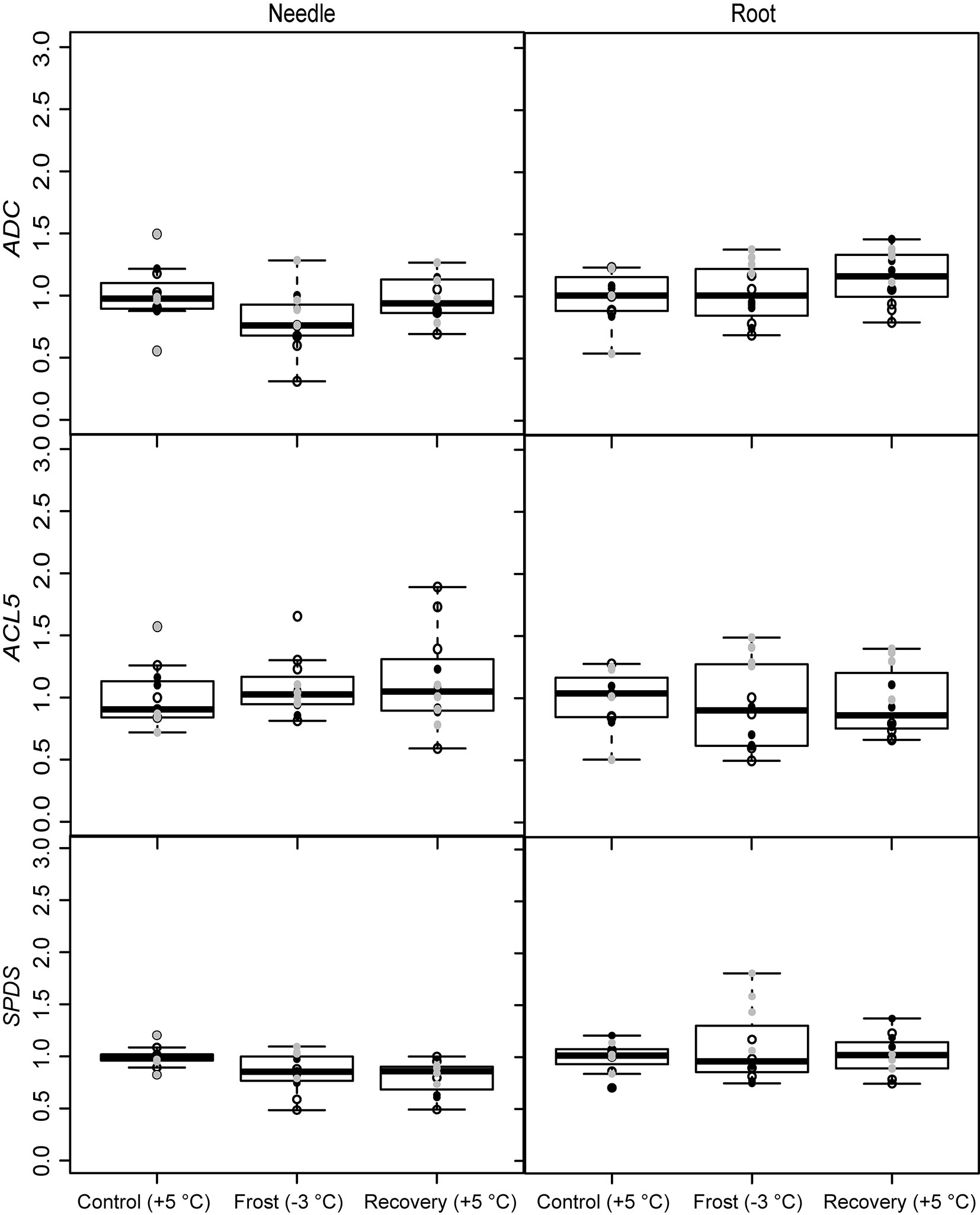
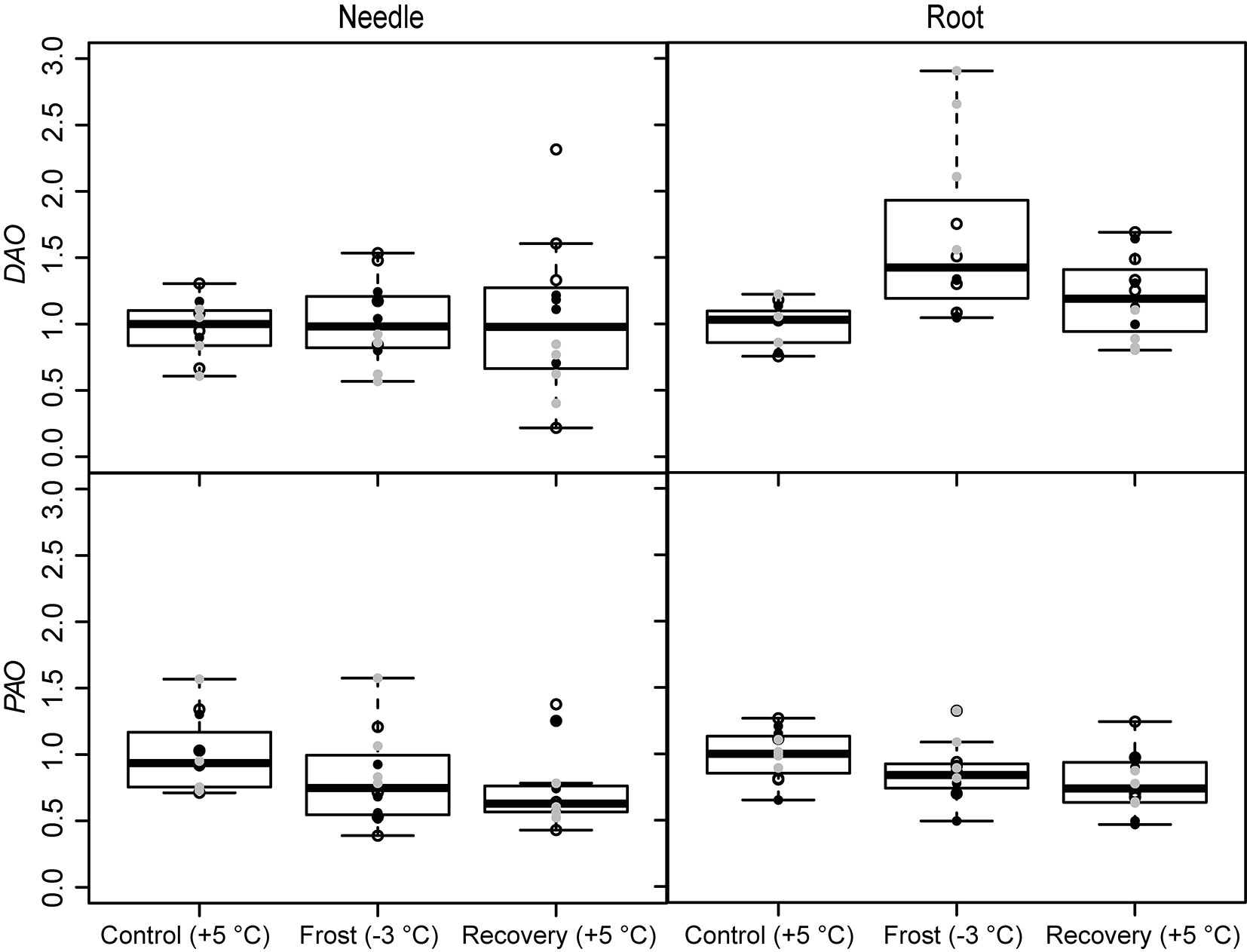
The expression of the PA metabolism related genes was affected by the frost and recovery treatments but differently in above and below seedling parts. The temperature change from +5 to -3 °C, decreased the expression of ADC and SPDS in needles. The expression of SPDS remained low level also after recovery (Fig. 3 and Tab. 2). Conversely, the expression of ADC increased in the roots after recovery. Frost increased also the expression of the PA catabolizing gene DAO in roots (Fig. 4 and Tab. 2), whereas the expression of PAO was down-regulated both in the needles and roots after recovery (Fig. 4 and Tab. 2). The levels of free or soluble conjugated PAs were not affected in the seedlings by the treatments (Tab. 3). However, the different families had different PA levels especially in their roots (Tab. 4).
Fig. 3 - Expression of the PA biosynthesizing genes (ADC, SPDS and ACL5) under spring frost (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) in Scots pine needles and roots. Open circles indicate the results from the family S1, black solid circles indicate the family S2 and grey circles indicate the family S3. Boxplot represents min and max values, first and third quartiles and mean value.
Tab. 2 - The effects of freezing stress treatments (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) on the gene expression results of needles and roots of Scots pine seedlings (linear model). Coefficients with 95% confidence intervals (CI) are presented. Coefficients with CIs not including zero are significant (indicated with “S”). See also Fig. 3 and Fig. 4. (a): difference between control and freezing stress; (b): difference between control and recovery treatment; (*): log transformation before model estimation.
| Needle | Control | Frost a | Recovery b | Root | Control | Frost a | Recovery b | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Est | 95% CI | Est | 95% CI | Est | 95% CI | Est | 95% CI | Est | 95% CI | Est | 95% CI | ||
| ADC | 1.00 | 0.86, 1.14 | -0.2s | -0.37, -0.04 | -0.03 | -0.19, 0.14 | ADC | 1 | 0.84, 1.16 | 0.03 | -0.12, 0.19 | 0.16s | 0.00, 0.31 |
| SPDS | 1.00 | 0.91, 1.09 | -0.15s | -0.27, -0.04 | -0.2s | -0.31, -0.08 | SPDS* | -0.01 | -0.14, 0.12 | 0.06 | -0.11, 0.23 | 0.02 | -0.15, 0.19 |
| ACL5* | -0.02 | -0.18, 0.13 | 0.08 | -0.12, 0.28 | 0.09 | -0.11, 0.29 | ACL5 | 1.00 | 0.74, 1.26 | -0.06 | -0.24, 0.12 | -0.04 | -0.22, 0.14 |
| DAO | 0.97 | 0.67, 1.27 | 0.05 | -0.23, 0.34 | 0.06 | -0.22, 0.34 | DAO* | -0.01 | -0.16, 0.14 | 0.45s | 0.24, 0.66 | 0.17 | -0.04, 0.38 |
| PAO* | -0.03 | -0.23, 0.16 | -0.25 | -0.52, 0.03 | -0.33s | -0.61, -0.05 | PAO | 1.00 | 0.88, 1.12 | -0.14 | -0.30, 0.02 | -0.22s | -0.38, -0.06 |
| SAMDC* | -0.13 | -0.59, 0.34 | -0.22 | -0.84, 0.39 | -0.44 | -1.05, 0.18 | SAMDC* | -0.18 | -0.49, 0.14 | 0.21 | -0.23, 0.65 | 0.13 | -0.13, 0.57 |
Fig. 4 - Expression of the PA catabolizing genes (DAO and PAO) under spring frost (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) in Scots pine needles and roots. Open circles indicate the results from the family S1, black solid circles indicate family S2 and grey circles indicate family S3. Boxplot represents min and max values, first and third quartiles and mean value.
Tab. 3 - The effects of freezing stress treatments (Control: +5 °C; Frost: -3 °C) on the free and soluble conjugated PA levels and phenol contents of needles and roots of Scots pine seedlings (linear model). Coefficients with 95% confidence intervals (CI) have been presented. Coefficients with CIs not including zero are significant. (a): difference between control and freezing stress; (b): difference between control and recovery treatment; (*): log transformation before model estimation.
| Needle | Control | Frost a | Root | Control | Frost a | ||||
|---|---|---|---|---|---|---|---|---|---|
| Est | 95% CI | Est | 95% CI | Est | 95% CI | Est | 95% CI | ||
| Free Put* | 5.62 | 5.39, 5.86 | -0.14 | -0.46, 0.17 | Free Put | 292.33 | 217.11, 367.56 | -6.27 | -56.74, 44.21 |
| Free Spd | 74.07 | 53.72, 94.42 | -12.73 | -41.51, 16.05 | Free Spd* | 4.05 | 3.62, 4.49 | 0.25 | -0.05, 0.54 |
| Free Spm* | 2.49 | 2.08, 2.91 | -0.05 | -0.64, 0.54 | Free Spm* | 2.59 | 2.06, 3.11 | 0.19 | -0.15, 0.54 |
| solc Put | 10.10 | 1.57, 18.62 | 7.65 | -4.41, 19.7 | solc Put | 42.94 | 13.06, 72.82 | -9.89 | -33.3, 13.52 |
| solc Spd | 28.09 | 11.78, 44.4 | -2.41 | -17.29, 12.48 | solc Spd | 26.22 | 8.50, 43.93 | -0.07 | -9.7, 9.56 |
| solc Spm | 6.60 | 1.15, 12.04 | -0.27 | -5.32, 4.78 | solc Spm | 5.96 | 0.93, 10.99 | -0.25 | -2.43, 1.94 |
| total phenols* | 0.34 | 0.24, 0.44 | 0.03 | -0.08, 0.14 | total phenols | 1.49 | 0.80, 2.18 | 0.23 | -0.16, 0.62 |
| pinosylvin* | -2.70 | -2.86, -2.54 | 0.12 | -0.11, 0.35 | pinosylvin | 0.04 | 0.03, 0.04 | 0 | -0.01, 0.01 |
| PSM* | -2.03 | -2.14, -1.93 | -0.01 | -0.13, 0.11 | PSM* | -1.88 | -2.04, -1.71 | 0.19 | -0.03, 0.42 |
Tab. 4 - Free and soluble conjugated PA contents (nmol g-1, FW) in the needles and roots of different Scots pine full sib families (S1, S2, S3). (Solc): Soluble conjugated.
| Portion | Family | Free Put | Free Spd | Free Spm | Solc Put | Solc Spd | Solc Spm | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | ||
| Needle | S1 | 307.9 | 517, 180 | 68.0 | 115, 6 | 13.5 | 21, 4 | 10.4 | 44.3, 0.0 | 12.2 | 41.1, 0.0 | 1.5 | 15.4, 0.0 |
| S2 | 314.0 | 857, 116 | 69.7 | 172, 23 | 18.1 | 61, 5 | 16.5 | 51.7, 0.0 | 32.5 | 76.7, 0.0 | 9.2 | 23.1, 0.0 | |
| S3 | 233.2 | 354, 111 | 65.4 | 131, 13 | 14.7 | 31, 1 | 14.9 | 49.3, 0.0 | 36.0 | 75.3, 17.4 | 8.7 | 21.3, 0.0 | |
| Root | S1 | 315.0 | 401, 197 | 75.1 | 144, 37 | 16.7 | 31, 8 | 17.8 | 50.9, 0.0 | 11.2 | 20.7, 0.0 | 1.3 | 5.0, 0.0 |
| S2 | 334.8 | 462, 198 | 95.1 | 142, 69 | 23.6 | 34, 11 | 31.3 | 20.7, 0.0 | 26.1 | 54.6, 7.9 | 6.3 | 12.8, 1.0 | |
| S3 | 217.8 | 313, 151 | 50.8 | 129, 24 | 10.9 | 24, 5 | 64.9 | 155.6, 18.2 | 41.3 | 63.7, 22.7 | 9.9 | 13.5, 3.3 | |
Phenylpropanoid and stilbene synthesis
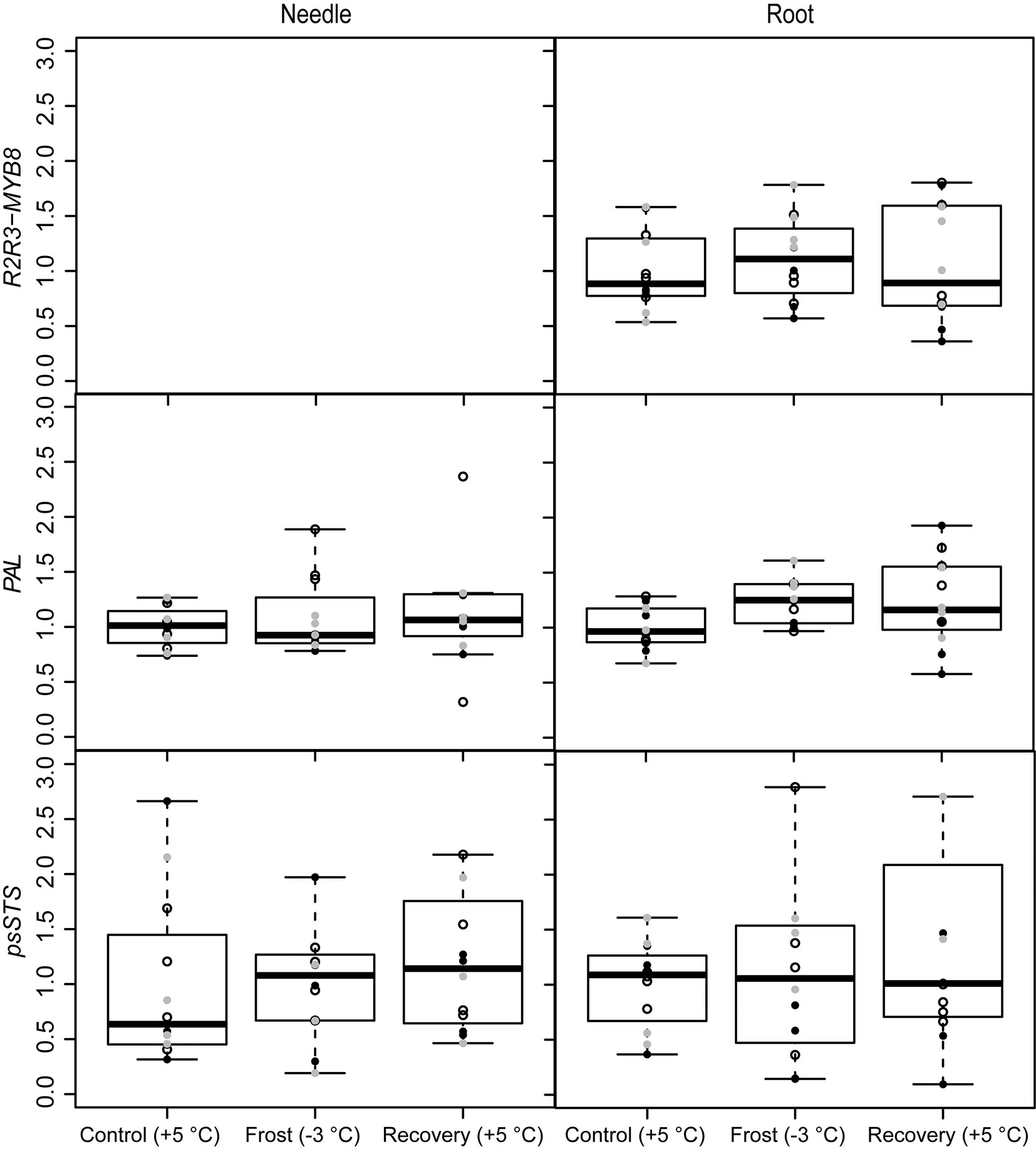
The R2R3-MYB8 gene was expressed only in roots but not in the needles (Fig. 5). The treatments did not affect the expression of R2R3-MYB8 in the roots (Tab. 1). Instead, the PAL gene was up-regulated in roots under frost (-3 °C) and after recovery (Fig. 5, Tab. 1). The pinosylvin synthetizing gene psSTS was not affected by treatments (Fig. 5, Tab. 1) and also levels of the stilbenes, i.e., pinosylvin and PSM, remained relatively stable during the experiment (Tab. 3). Phenol contents were on the same level in all three families in needles and roots (Tab. 5).
Fig. 5 - Expression of the phenylpropanoid synthesis related genes (R2R3-MYB8 and PAL) and pinosylvin synthase (psSTS) under spring frost (Control: +5 °C; Frost: -3 °C; Recovery: +5 °C) in Scots pine needles and roots. Open circles indicate the results from the family S1, black solid circles indicate family S2 and grey circles family S3. Boxplot represents min and max values, first and third quartiles and mean value.
Tab. 5 - Pinosylvin, PSM (nmol g-1, FW) and total phenol (mg g-1, FW) content in needles and roots of Scots pine full sib families (S1, S2, S3).
| Portion | Family | Pinosylvin | PSM | Total phenols | |||
|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | ||
| Needle | S1 | 0.08 | 0.20, 0.05 | 0.14 | 0.19, 0.11 | 1.51 | 1.75, 1.27 |
| S2 | 0.07 | 0.08, 0.06 | 0.13 | 0.14, 0.11 | 1.32 | 1.48, 1.20 | |
| S3 | 0.07 | 0.10, 0.06 | 0.12 | 0.15, 0.09 | 1.50 | 2.05, 1.14 | |
| Root | S1 | 0.03 | 0.05, 0.02 | 0.18 | 0.36, 0.11 | 2.27 | 3.06, 1.74 |
| S2 | 0.04 | 0.05, 0.03 | 0.19 | 0.37, 0.13 | 1.36 | 2.09, 0.91 | |
| S3 | 0.04 | 0.05, 0.03 | 0.16 | 0.20, 0.12 | 1.20 | 2.00, 0.21 | |
Discussion
Late winter and spring weather conditions have great significance on the population dynamics of Scots pine at the northern timberline ([21]). Humidity and temperature affect the timing of Scots pine seed germination. After winter, temperatures have to be continuously over 10 degrees, before seeds start to germinate ([32]). In southern Finland this usually occurs in the middle of May, although seedlings frequently face below zero conditions during the early summer nights. The risk of this frost damage occurrence is predicted to increase due to climate change in the future. PA biosynthesis with transcriptional and metabolic changes occurs in response to such low temperatures in many plant species ([1]). However, there are no other studies where PA metabolism, phenylpropanoid synthesis and freezing stress have been simultaneously investigated in Pinus species.
Generally, cold stress can have adverse effects on plant growth and development ([6]). Cold acclimation usually involves regulation at transcriptional, post-transcriptional and post-translational levels ([29], [45]). The results of our present spring frost study suggest that PA metabolism is down-regulated in Scots pine needles when the temperature falls below zero. Spring frost decreased expression of ADC, SPDS, ICE1 and LEA and the lower expression level of SPDS and ICE1 was maintained after the recovery in the needles. In our previous studies, we have shown that under drought or osmotic stresses various tissues are able to maintain consistent PA levels due to a down-regulation PA catabolizing genes, which seems to be advantageous ([30], [31]). In the present study, the expression of PAO decreased in needles and roots after the recovery, which might be a consequence of the frost induced down-regulation of the PA biosynthesizing genes ADC and SPDS. ICE1 is a transcription factor constitutively expressed in plant cells and the post-translational modifications are necessary for ICE1 protein to activate downstream genes ([6]). Therefore, the transcription level down-regulation of ICE1 under frost might suggest general metabolic rigor in needle tissues rather than the regulation of freezing stress induced cascades in the cells. However, transcription level gene regulation differed between above and below ground parts of the seedlings.
Spring frost induced expression of the PA catabolizing gene DAO in Scots pine roots. Analogously, freezing stress increased DAO and PAO activity in Virginia pine (Pinus virginiana Mill.) roots and exogenously applied PAs increased root growth at low temperatures ([44]). A soil temperature of +5 °C inhibits the growth of roots in Scots pine seedlings, leading to decreased ATP enzyme activity and an imbalance between water and nutrient uptake and transpiration ([37]). In the present study, increased DAO expression in roots may indicate the launching of a signaling pathway where PA catabolism is up-regulated and H2O2 produced, leading to alterations in root development. Expression of the H2O2-scavenging enzyme gene CAT remained stable, which is consistent with our earlier results, suggesting that changes in CAT expression are not necessarily connected with stress in developing Scots pine tissues. Rather, CAT seems to protect cells against H2O2 accumulation, especially during active metabolism ([48]).
An increase of PA levels appears to be necessary for the roots of Virginia pine to resist cold stress ([44]). PAs conjugate with hydroxycinnamic acids, proteins and cell wall components and are shown to stabilize membranes under abiotic stress ([2]). Accordingly, Put was required for root growth in Virginia pine plantlets, whereas Spd and Spm behave as growth inhibitors ([44]). Experimental, spring frost conditions did not affect free or soluble conjugated PA levels in Scots pine seedlings. In general, tolerant plant species seem to accumulate PAs under freezing stress more than the non-tolerant ones ([13]).
PA oxidation has been associated with stress-induced phenylpropanoid metabolism in cell cultures of the grape vine (Vitis vinifera L. - [17]). PA oxidation releases H2O2, which promotes cell-wall stiffening through formation of covalent cross links between cell wall components ([34]). R2R3-MYB genes are transcription factors belonging to a large gene family and known to regulate phenylpropanoid synthesis related genes such as PAL and 4-coumarate:CoA ligase (4CL - [8]). Different R2R3-MYB genes have a dual action in regulating their response genes ([4]). Plants can alter their metabolism in response to environmental cues via the R2R3-MYB converse, reflecting phenotypic plasticity ([4]). In maritime pine seedlings, phenylpropanoid synthesis related genes were induced by a high expression of R2R3-MYB8 in lignifying tissues ([8]). In spruce, the R2R3-MYB8 gene expressed in differentiating xylem in roots and stems, having a very similar expression profile with 4CL ([4]). In the present study, the putative R2R3-MYB8 gene of Scots pine was mainly expressed in the roots, but only very mildly in needles. Expression of the PAL gene increased in roots under freezing stress and remained high throughout the recovery treatment, suggesting a frost -induced activation of the phenylpropanoid pathway in roots.
In different microhabitats (e.g., in peatland soil) the seedlings are exposed to rapid changes in temperature, especially in spring. Higher PA or phenolic contents might be advantageous to seedlings under such conditions, which do not allow for prolonged cellular metabolic responses. Jensen et al. ([18]) found that PAL activity as well as the total amount of phenolics increased in root tissue 24 h after soybean (Glycine max [L.] Merr) plants were transferred to low temperatures. PAL activity, and the accumulation of soluble phenolics, hydroxyl cinnamic acids and anthocyanins depends on the temperature to which the plants are exposed, particularly so in chilling resistant plants ([42]). In oil plants (Brassica napus L.), decrease in content of ferulic acid in the cell walls of PAL-inhibited cells resulted in cell wall rigidity and cold stress sensitivity ([42]). Therefore, the results of the present study suggest that the transcriptional up-regulation of the PA catabolizing gene DAO and phenylpropanoid synthesizing gene PAL can be connected to an early response of root development in Scots pine seedlings via root growth inhibition and/or cell-wall stiffening processes.
In our earlier study we have shown that under severe drought stress Scots pine seedlings invest in the shoots rather than roots ([30]). This is the case also after freezing stress as rising temperatures decreases the root/shoot ratio of Scots pine seedlings, apparently due to a preferential allocation of photosynthates to shoots ([46]). In the present study, the transcription level data shows that gene expressions remained stable or decreased in needles under slight freezing stress. Conversely, DAO and PAL genes were up-regulated in roots under subzero temperatures suggesting that spring frost induced signaling pathway leads to reorganization of root growth in Scots pine seedlings.
Conclusions
An increased expression of DAO in roots under freezing stress and an up-regulation of ADC after recovery treatment suggest that PA metabolism has a specific role in spring frost-induced signaling in the root tissue. Moreover, up-regulation of the PAL gene in roots due to freezing may be associated with the initiation of phenylpropanoid synthesis, although this could not be sustained via changes in PA or phenol contents during the present experiment. This is the first time that PA oxidation and phenylpropanoid biosynthesis related genes have been shown to coexpress under mild freezing stress, mimicking spring frost in the root tissue of pine seedlings. This reveals their potential role in the initial stages of the coping strategies of Scots pine against spring frost.
List of abbreviations
The following abbreviations were used throughout the text:
- ACL5: thermospermine synthase;
- ADC: arginine decarboxylase;
- CaPF1: freezing tolerance related protein 1 of pepper;
- CAT: catalase;
- CBF: C-repeat binding factor;
- CI: confidence interval;
- DAO: diamine oxidase;
- HCAA: hydroxycinnamic acid amide;
- H2O2: hydrogen peroxide;
- HPLC: high pressure liquid chromatography;
- ICE1: freezing stress related gene;
- LEA: late-embryogenesis-abundant protein;
- PA: polyamine;
- PAL: phenylalanine lyase;
- PAO: polyamine oxidase;
- PMT: pinosylvin monomethyl transferase;
- PSM: pinosylvin monomethyl ether;
- Put: putrescine;
- R2R3-MYB: transcription factor family related to phenylpropanoid synthesis;
- SAMS: S-adenosyl methionine synhase;
- SAMDC: S-adenosyl methionine decarboxylase;
- Spd: spermidine;
- SPDS: spermidine synthase;
- Spm: spermine;
- SPMS: spermine synthase;
- psSTS: pinosylvin synthase;
- Tspm: thermospermine;
- TUBA: alfa-tubulin;
- UBQ: ubiquitin;
- 4CL: 4-coumarate:CoA ligase.
Acknowledgements
RMM designed the practical part of the study, carried out the molecular genetic studies, analyzed the data and wrote the manuscript. LH performed the statistical analyses. MS and TS helped to design and arrange the experimental platform. HH conceived of the study and together with TS and JV participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
We are grateful to Ms. Eeva Pihlajaviita, Ms. Anneli Käenmäki and Ms. Hanna Leppälammi for their skillful technical help. We thank language editor Dr. Otso Huitu who revised English of the manuscript. The research was funded by the Thule Institute (2010-2013) (to HH), Academy of Finland (Project 121994 to TS) and Graduate School of Forest Sciences (2012-2016) (to RMM).
References
CrossRef | Gscholar
CrossRef | Gscholar
CrossRef | Gscholar
Gscholar
CrossRef | Gscholar
Gscholar
CrossRef | Gscholar
Gscholar
Gscholar
Authors’ Info
Authors’ Affiliation
Markku Saarinen
Tytti Sarjala
Natural Resources Institute Finland (Luke), Parkano Research Unit, FI-39700 Parkano (Finland)
Jaana Vuosku
Hely Häggman
Genetics and Physiology department, University of Oulu, P.O. Box 3000, FI-90014 Oulu (Finland)
Natural Resources Institute Finland (Luke), Vantaa Research Unit, FI- 01370, Vantaa (Finland)
Natural Resources Institute Finland (Luke), Punkaharju Research Unit, FI-58450, Punkaharju (Finland)
Corresponding author
Paper Info
Citation
Muilu-Mäkelä R, Vuosku J, Saarinen M, Hamberg L, Ruotsalainen S, Häggman H, Sarjala T (2017). Coping with spring frost-effects on polyamine metabolism of Scots pine seedlings. iForest 10: 227-236. - doi: 10.3832/ifor2003-009
Academic Editor
Mike Perks
Paper history
Received: Feb 05, 2016
Accepted: Nov 27, 2016
First online: Jan 27, 2017
Publication Date: Feb 28, 2017
Publication Time: 2.03 months
Copyright Information
© SISEF - The Italian Society of Silviculture and Forest Ecology 2017
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Web Metrics
Breakdown by View Type
Article Usage
Total Article Views: 50634
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 42338
Abstract Page Views: 3008
PDF Downloads: 3922
Citation/Reference Downloads: 36
XML Downloads: 1330
Web Metrics
Days since publication: 3322
Overall contacts: 50634
Avg. contacts per week: 106.69
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2017): 3
Average cites per year: 0.33
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
Related Contents
iForest Similar Articles
Research Articles
Links between phenology and ecophysiology in a European beech forest
vol. 8, pp. 438-447 (online: 15 December 2014)
Research Articles
Relationship between frost hardiness of adults and seedlings of different tree species
vol. 7, pp. 282-288 (online: 01 April 2014)
Research Articles
Growing season water balance of an inner alpine Scots pine (Pinus sylvestris L.) forest
vol. 11, pp. 469-475 (online: 02 July 2018)
Review Papers
Drought-induced mortality of Scots pines at the southern limits of its distribution in Europe: causes and consequences
vol. 3, pp. 95-97 (online: 15 July 2010)
Research Articles
Sap flow, leaf-level gas exchange and spectral responses to drought in Pinus sylvestris, Pinus pinea and Pinus halepensis
vol. 10, pp. 204-214 (online: 01 November 2016)
Research Articles
The growth dynamics of East European Scots pine (Pinus sylvestris L.) populations - a Lithuanian field trial
vol. 17, pp. 59-68 (online: 06 March 2024)
Research Articles
Wintertime photosynthesis and spring recovery of Ilex aquifolium L.
vol. 12, pp. 389-396 (online: 31 July 2019)
Research Articles
Identification and allelochemical activity of phenolic compounds in extracts from the dominant plant species established in clear-cuts of Scots pine stands
vol. 10, pp. 309-314 (online: 23 February 2017)
Research Articles
Growth patterns of Scots pine (Pinus sylvestris L.) under the current regional pollution load in Lithuania
vol. 8, pp. 509-516 (online: 12 November 2014)
Short Communications
Preliminary indications for diverging heat and drought sensitivities in Norway spruce and Scots pine in Central Europe
vol. 13, pp. 89-91 (online: 01 March 2020)
iForest Database Search
Search By Author
Search By Keyword
Google Scholar Search
Citing Articles
Search By Author
Search By Keywords
PubMed Search
Search By Author
Search By Keyword