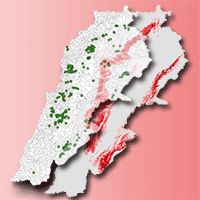
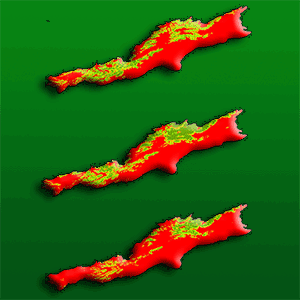
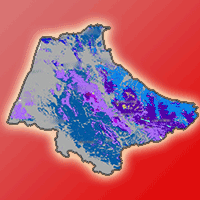
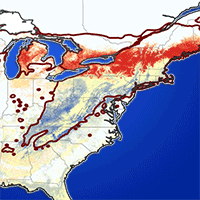
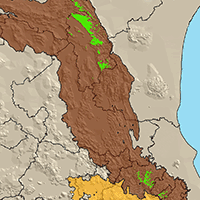
The different models that predict the distribution of species are a useful tool for the evaluation and monitoring of forest resources as they facilitate the planning of their management in a changing climate environment. Recently, a significant number of algorithms have been proposed for this purpose, making it difficult to select the most appropriate to use. The evaluation of performance and predictive stability of these models can elucidate this problem. Distribution data of 17 pine species with high economic importance for Mexico were collected and distribution models were carried out. We carried out a pre-modeling design to select the prediction variables (climatic, edaphic and topographic), after which nine algorithms and an ensemble model were contrasted against one another. The true skill statistic (TSS) and the area under the curve (AUC) were used to evaluate the predictive performance of the models, and the coefficient of variation of the predictions was used to evaluate their stability. The number of predictive variables in the final models fluctuated from 6 to 12; the mean diurnal range and the maximum temperature of warmest month were included in the models for most species. Random forests, the ensemble model, generalized additive models and MaxEnt were the ones that best described the distribution of the species (AUC >0.92 and TSS >0.72); the opposite was found in Bioclim and Domain (AUC<0.75 and <0.82; and TSS<0.5 and <0.55). Support vector machine, Mahalanobis distance, generalized linear models and boosted regression trees obtained intermediate settings. The coefficient of variation indicated that Bioclim, Domain and Support vector machine have low predictive stability (CV>0.055); on the contrary, Maxent and the ensemble model attained high predictive stability (CV<0.015). The ensemble model obtained greater performance and predictive stability in the predictions of the distribution of the 17 species of pines. The differences found in performance and predictive stability of the algorithms suggest that the ensemble model has the potential to model the distribution of tree species.
Keywords
, , , , , , ,
Citation
Montoya-Jiménez JC, Valdez-Lazalde JR, Ángeles-Perez G, De Los Santos-Posadas HM, Cruz-Cárdenas G (2022). Predictive capacity of nine algorithms and an ensemble model to determine the geographic distribution of tree species. iForest 15: 363-371. - doi: 10.3832/ifor4084-015
Academic Editor
Maurizio Marchi
Paper history
Received: Feb 22, 2022
Accepted: Jul 12, 2022
First online: Sep 20, 2022
Publication Date: Oct 31, 2022
Publication Time: 2.33 months
© SISEF - The Italian Society of Silviculture and Forest Ecology 2022
Open Access
This article is distributed under the terms of the Creative Commons Attribution-Non Commercial 4.0 International (https://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.

Breakdown by View Type
(Waiting for server response...)
Article Usage
Total Article Views: 32916
(from publication date up to now)
Breakdown by View Type
HTML Page Views: 27544
Abstract Page Views: 2933
PDF Downloads: 1953
Citation/Reference Downloads: 17
XML Downloads: 469
Web Metrics
Days since publication: 1238
Overall contacts: 32916
Avg. contacts per week: 186.12
Article Citations
Article citations are based on data periodically collected from the Clarivate Web of Science web site
(last update: Mar 2025)
Total number of cites (since 2022): 12
Average cites per year: 3.00
Publication Metrics
by Dimensions ©
Articles citing this article
List of the papers citing this article based on CrossRef Cited-by.
(1)
Aceves-Rangel LD, Méndez-González J, García-Aranda MA, Nájera-Luna JA (2018)Distribución potencial de 20 especies de pinos en México [Potential distribution of 20 pine species in Mexico]. Agrociencia 52: 1043-1057. [in Spanish]
Gscholar
(2)
Araújo MB, New M (2007)Ensemble forecasting of species distributions. Trends in Ecology and Evolution 22: 42-47.
CrossRef |
Gscholar
(3)
Betancourt GA (2005)Las máquinas de soporte vectorial (SVMs) [Vector support machines (SVMs)]. Scientia et Technica 11 (27): 67-72. [in Spanish]
Online |
Gscholar
(4)
Bivand R, Keitt T, Rowlingson B, Pebesma E, Sumner M, Hijmans R, Rouault E, Warmerdam F, Ooms J, Rundel C (2020)Package “rgdal”. R package version 1:5-18.
Online |
Gscholar
(5)
Carpenter G, Gillison AN, Winter J (1993)DOMAIN: a flexible modelling procedure for mapping potential distributions of plants and animals. Biodiversity and Conservation 2: 667-680.
CrossRef |
Gscholar
(6)
Chapman AD (2005)Principles and methods of data cleaning - Primary species and species-occurrence data (version 1.0). Report for the Global Biodiversity Information Facility - GBIF, Copenhagen, Denmark, pp. 72.
Gscholar
(7)
Cobos ME, Peterson AT, Osorio-Olvera L, Jiménez-García D (2019)An exhaustive analysis of heuristic methods for variable selection in ecological niche modeling and species distribution modeling. Ecological Informatics 53: 100983.
CrossRef |
Gscholar
(8)
CONABIO (2020)Sistema Nacional de Información sobre Biodiversidad. Registros de ejemplares [National Information System on Biodiversity. Specimen Records]. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO), Ciudad de México, México, web site. [in Spanish]
Online |
Gscholar
(9)
CONAFOR (2019)Estado que guarda el sector forestal en México [Status of the Forest Sector in Mexico]. Comisión Nacional Forestal (CONAFOR), Zapopan Jalisco, México, pp. 406. [in Spanish].
Online |
Gscholar
(10)
Conrad O, Bechtel B, Bock M, Dietrich H, Fischer E, Gerlitz L, Wehberg J, Wichmann V, Böhner J (2015)System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geoscientific Model Development 8: 1991-2007.
CrossRef |
Gscholar
(11)
Elith J, H. Graham C, P. Anderson R, Dudík M, Ferrier S, Guisan A, J. Hijmans R, Huettmann F, R. Leathwick J, Lehmann A, Li J, G. Lohmann L, A. Loiselle B, Manion G, Moritz C, Nakamura M, Nakazawa Y, Mcc. M. Overton J, Townsend Peterson A, J. Phillips S, Richardson K, Scachetti-Pereira R, E. Schapire R, Soberón J, Williams S, S. Wisz M, E. Zimmermann N (2006)Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29 (2): 129-151.
CrossRef |
Gscholar
(12)
Escobar LE, Lira-Noriega A, Medina-Vogel G, Peterson AT (2014)Potential for spread of the white-nose fungus (
Pseudogymnoascus destructans) in the Americas: use of Maxent and NicheA to assure strict model transference. Geospatial Health 9: 221-229.
CrossRef |
Gscholar
(13)
Etherington TR (2019)Mahalanobis distances and ecological niche modelling: correcting a chi-squared probability error. PeerJ 7: 1-8.
CrossRef |
Gscholar
(14)
Farjon A, Filer D (2013)An atlas of the world’s conifers. Brill, Leiden, The Netherlands, pp. 512.
CrossRef |
Gscholar
(15)
Fick SE, Hijmans RJ (2017)WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302-4315.
CrossRef |
Gscholar
(16)
García-Aranda MA, Méndez-González J, Hernández-Arizmendi JY (2018)Distribución potencial de
Pinus cembroides, Pinus nelsonii y
Pinus culminicola en el Noreste de México [Potential distribution of
Pinus cembroides, Pinus nelsonii and
Pinus culminicola in northeastern Mexico]. Ecosistemas y Recursos Agropecuarios 5: 3-13. [in Spanish]
CrossRef |
Gscholar
(17)
GBIF (2020)Occurrence download. Global Biodiversity Information Facility, Copenhagen, Denmark, web site.
Online |
Gscholar
(18)
Guisan A, Edwards TC, Hastie T (2002)Generalized linear and generalized additive models in studies of species distributions: setting the scene. Ecological Modelling 157: 89-100.
CrossRef |
Gscholar
(19)
Hansen MC, Defries RS, Townshend JRG, Sohlberg R (2000)Global land cover classification at 1 km spatial resolution using a classification tree approach. International Journal of Remote Sensing 21: 1331-1364.
CrossRef |
Gscholar
(20)
Hao T, Elith J, Guillera-Arroita G, Lahoz-Monfort JJ (2019)A review of evidence about use and performance of species distribution modelling ensembles like BIOMOD. Diversity and Distributions 25: 839-852.
CrossRef |
Gscholar
(21)
Hao T, Elith J, Lahoz-Monfort JJ, Guillera-Arroita G (2020)Testing whether ensemble modelling is advantageous for maximising predictive performance of species distribution models. Ecography 43: 549-558.
CrossRef |
Gscholar
(22)
Hengl T, Mendes De Jesus J, Heuvelink GBM, Ruiperez Gonzalez M, Kilibarda M, Blagotić A, Shangguan W, Wright MN, Geng X, Bauer-Marschallinger B, Guevara MA, Vargas R, MacMillan RA, Batjes NH, Leenaars JGB, Ribeiro E, Wheeler I, Mantel S, Kempen B (2017)SoilGrids250m: global gridded soil information based on machine learning. PLoS One 12: 1-40.
CrossRef |
Gscholar
(23)
Hijmans RJ, Guarino L, Bussink C, Mathur P, Cruz M, Barrantes I, Rojas E (2012)DIVA-GIS, ver. 7.5. A geographic information system for the analysis of species distribution data. Versão 7: 476-486.
Gscholar
(24)
Hijmans RJ, Elith J (2017)Species distribution modeling with R. Web site.
Online |
Gscholar
(25)
Hijmans RJ, Etten Van J, Sumner M, Cheng J, Bevan A, Bevan R, Busetto L, Canty M, Forrest D, Ghosh A, Golicher D, Gray J, Greenberg JA (2020)Package “raster”. R Package version 3:3-13.
Online |
Gscholar
(26)
INE/INEGI (1997)Conjunto nacional de uso del suelo y vegetación a escala 1:250.000, Serie I [National collection of land use and vegetation scale 1:250.000, Series I]. DOEG-INEGI, México City, Mexico. [in Spanish]
Gscholar
(27)
INEGI (2016)Conjunto nacional de uso del suelo y vegetación a escala 1:250.000, Serie VI [National collection of land use and vegetation scale 1:250.000, Series VI]. Instituto Nacional de Estadística y Geografía (INEGI), México City, Mexico. [in Spanish]
Gscholar
(28)
Jarnevich CS, Young NE (2019)Not so normal normals: species distribution model results are sensitive to choice of climate normals and model type. Climate 7: 1-15.
CrossRef |
Gscholar
(29)
Manzanilla-Quiñones U, Aguirre-Calderón OA, Jiménez-Pérez J, Treviño-Garza EJ, Yerena-Yamallel JI (2019)Distribución actual y futura del bosque subalpino de
Pinus hartwegii Lindl en el Eje Neovolcánico Transversal [Current and future distribution of the
Pinus hartwegii Lindl subalpine forest in the Tranverse Neovolcanic Belt]. Madera y Bosques 25: 1-16. [in Spanish]
CrossRef |
Gscholar
(30)
Marchi M, Ducci F (2018)Some refinements on species distribution models using tree-level national forest inventories for supporting forest management and marginal forest population detection. iForest 11: 291-299.
CrossRef |
Gscholar
(31)
Marmion M, Parviainen M, Luoto M, Heikkinen RK, Thuiller W (2009)Evaluation of consensus methods in predictive species distribution modelling. Diversity and Distributions 15: 59-69.
CrossRef |
Gscholar
(32)
Mi C, Huettmann F, Guo Y, Han X, Wen L (2017)Why choose Random Forest to predict rare species distribution with few samples in large undersampled areas? Three Asian crane species models provide supporting evidence. PeerJ 5: 1-22.
CrossRef |
Gscholar
(33)
Naimi B, Araújo MB (2016)SDM: a reproducible and extensible R platform for species distribution modelling. Ecography 39: 368-375.
CrossRef |
Gscholar
(34)
Nix HA (1986)A biogeographic analysis of Australian elapid snakes. In: “Atlas of elapid snakes of Australia” (Longmore R ed). Australian Flora and Fauna Series 7, Bureau of Flora and Fauna, Canberra, Australia, pp. 4-15.
Gscholar
(35)
Osorio-Olvera L, Lira-Noriega A, Soberón J, Peterson AT, Falconi M, Contreras-Díaz RG, Martínez-Meyer E, Barve V, Barve N (2020)ntbox: an R package with graphical user interface for modelling and evaluating multidimensional ecological niches. Methods in Ecology and Evolution 11: 1199-1206.
CrossRef |
Gscholar
(36)
Pecchi M, Marchi M, Moriondo M, Forzieri G, Ammoniaci M, Bernetti I, Bindi M, Chirici G (2020)Potential impact of climate change on the spatial distribution of key forest tree species in Italy under RCP4.5 for 2050s. Forest 11: 1-19.
CrossRef |
Gscholar
(37)
Pecchi M, Marchi M, Burton V, Giannetti F, Moriondo M, Bernetti I, Bindi M, Chirici G (2019)Species distribution modelling to support forest management. A literature review. Ecological Modelling 411: 108817.
CrossRef |
Gscholar
(38)
Perry JP (1991)The pines of Mexico and Central America. Timber Press, Portland, OR, USA, pp. 231.
Online |
Gscholar
(39)
Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB (2012)Ecological niches and geographic distributions (MPB-49). Series “Monographs in Population Biology”, vol. 49, Princeton University Press, Princeton, NJ, USA, pp. 316.
CrossRef |
Gscholar
(40)
Phillips SJ, Anderson RP, Schapire RE (2006)Maximum entropy modeling of species geographic distributions. Ecological Modelling 190: 231-259.
CrossRef |
Gscholar
(41)
R Core Team (2020)R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Online |
Gscholar
(42)
Radosavljevic A, Anderson RP (2014)Making better Maxent models of species distributions: complexity, overfitting and evaluation. Journal of Biogeography 41: 629-643.
CrossRef |
Gscholar
(43)
Ramos-Dorantes DB, Villaseñor JL, Ortiz E, Gernandt DS (2017)Biodiversity, distribution, and conservation status of Pinaceae in Puebla, Mexico. Revista Mexicana de Biodiversidad 88: 215-223.
CrossRef |
Gscholar
(44)
Ren-Yan D, Xiao-Quan K, Min-Yi H, Wei-Yi F, Zhi-Gao W (2014)The predictive performance and stability of six species distribution models. PLoS One 9.
CrossRef |
Gscholar
(45)
Reynoso Santos R, Pérez Hernández MJ, López Baez W, Hernández Ramos J, Muñoz Flores HJ, Cob Uicab JV, Reynoso Santos MD (2018)El nicho ecológico como herramienta para predecir áreas potenciales de dos especies de pino [The ecological niche as a tool for predicting potential areas of two pine species]. Revista Mexicana de Ciencias Forestales 9: 47-68. [in Spanish]
CrossRef |
Gscholar
(46)
Shabani F, Kumar L, Ahmadi M (2016)A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecology and Evolution 6: 5973-5986.
CrossRef |
Gscholar
(47)
Soberón J, Osorio-Olvera L, Peterson T (2017)Diferencias conceptuales entre modelación de nichos y modelación de áreas de distribución [Conceptual differences between ecological niche modeling and species distribution modeling]. Revista Mexicana de Biodiversidad 88: 437-441. [in Spanish]
CrossRef |
Gscholar
(48)
Vargas-Larreta B, Corral-Rivas JJ, Aguirre-Calderón OA, López-Martínez JO, De los Santos-Posadas HM, Zamudio-Sánchez FJ, Treviño-Garza EJ, Martínez-Salvador M, Aguirre-Calderón CG (2017)SiBiFor: forest biometric system for forest management in Mexico. Revista Chapingo Serie Ciencias Forestales y Del Ambiente 23: 437-455.
CrossRef |
Gscholar
(49)
Velazco SJE, Galvão F, Villalobos F, De Marco Júnior P (2017)Using worldwide edaphic data to model plant species niches: An assessment at a continental extent. PLoS One 12: 1-24.
CrossRef |
Gscholar
(50)
Watling JI, Romañach SS, Bucklin DN, Speroterra C, Brandt LA, Pearlstine LG, Mazzotti FJ (2012)Do bioclimate variables improve performance of climate envelope models? Ecological Modelling 246: 79-85.
CrossRef |
Gscholar